
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,893,230 – 9,893,324 |
| Length | 94 |
| Max. P | 0.996678 |

| Location | 9,893,230 – 9,893,324 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 72.76 |
| Shannon entropy | 0.54566 |
| G+C content | 0.58398 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -22.55 |
| Energy contribution | -25.04 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.996678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
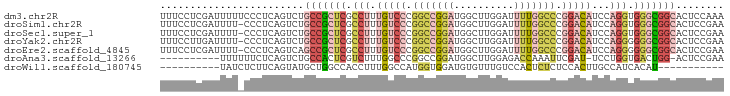
>dm3.chr2R 9893230 94 + 21146708 UUUCCUCGAUUUUUCCCUCAGUCUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCAAA .......((((........))))((((((((((((.(((((.(((((((..........))))))).)))))...))))))))))))....... ( -42.00, z-score = -3.20, R) >droSim1.chr2R 8329766 93 + 19596830 UUUCCUCGAUUUU-CCCUCAGUCUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCGAA .............-...((.(..((((((((((((.(((((.(((((((..........))))))).)))))...))))))))))))..).)). ( -43.10, z-score = -3.20, R) >droSec1.super_1 7418377 93 + 14215200 UUUCCUCGAUUUU-CCCUCAGUCUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCGAA .............-...((.(..((((((((((((.(((((.(((((((..........))))))).)))))...))))))))))))..).)). ( -43.10, z-score = -3.20, R) >droYak2.chr2R 14078698 93 - 21139217 UUUCCUUGAUUUU-CCCUCAGUCUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGGGGGCGGCACUCCGAA .............-...((.(..((((((((.(((.(((((.(((((((..........))))))).)))))...))).))))))))..).)). ( -40.90, z-score = -1.89, R) >droEre2.scaffold_4845 6660634 93 - 22589142 UUUCCUCGAUUUU-CCCUCAGUCAGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGGGGGCGGCACUCCGAA .....((((((..-.....)))).(((((((.(((.(((((.(((((((..........))))))).)))))...))).))))))).....)). ( -39.90, z-score = -1.39, R) >droAna3.scaffold_13266 13877511 82 - 19884421 ----------UUUUUUCUCAGUCUGCCACUCGUCUUUGGCCCGGCCGGAUGGCUUGGAGACCAAAUUCGAU-UCCUGGUGACUGG-ACUCCGAA ----------.....((.(((((.((((.(((..((((((((((((....))).))).).)))))..))).-...))))))))))-)....... ( -27.20, z-score = -1.42, R) >droWil1.scaffold_180745 1099893 73 - 2843958 ----------UAUCUCUUCAGUAUGCUGGCCACCUUUGGCCAUGGUGGAUGUGUUUGUCCACUCUCUCCACUUGCCAUCACAU----------- ----------..........((((((((((((....)))))).(((((((......)))))))..........).))).))..----------- ( -18.60, z-score = -1.12, R) >consensus UUUCCUCGAUUUU_CCCUCAGUCUGCCGCUCGCCUUUGUCCCGGCCGGAUGGCUUGGAUUUUGGCCCGGACAUCCAGGUGGGCGGCACUCCGAA ........................(((((((.(((.(((((.(((((((..........))))))).)))))...))).)))))))........ (-22.55 = -25.04 + 2.50)

| Location | 9,893,230 – 9,893,324 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 72.76 |
| Shannon entropy | 0.54566 |
| G+C content | 0.58398 |
| Mean single sequence MFE | -33.49 |
| Consensus MFE | -19.71 |
| Energy contribution | -23.15 |
| Covariance contribution | 3.43 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
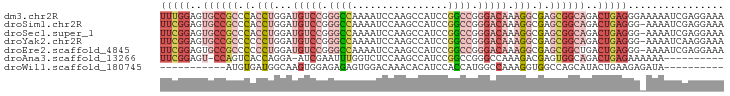
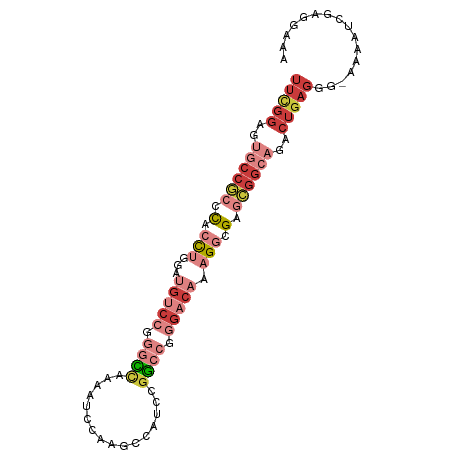
>dm3.chr2R 9893230 94 - 21146708 UUUGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAGACUGAGGGAAAAAUCGAGGAAA ((..(..((((((.(.(((...(((((.((((................)))).))))).))).).))))))..)..))................ ( -35.99, z-score = -1.21, R) >droSim1.chr2R 8329766 93 - 19596830 UUCGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAGACUGAGGG-AAAAUCGAGGAAA (((((..((((((.(.(((...(((((.((((................)))).))))).))).).))))))..)))))..-............. ( -38.39, z-score = -1.74, R) >droSec1.super_1 7418377 93 - 14215200 UUCGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAGACUGAGGG-AAAAUCGAGGAAA (((((..((((((.(.(((...(((((.((((................)))).))))).))).).))))))..)))))..-............. ( -38.39, z-score = -1.74, R) >droYak2.chr2R 14078698 93 + 21139217 UUCGGAGUGCCGCCCCCCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAGACUGAGGG-AAAAUCAAGGAAA (((((..((((((.(.(((...(((((.((((................)))).))))).))).).))))))..)))))..-............. ( -37.49, z-score = -1.24, R) >droEre2.scaffold_4845 6660634 93 + 22589142 UUCGGAGUGCCGCCCCCCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCUGACUGAGGG-AAAAUCGAGGAAA (((((...(((((.(.(((...(((((.((((................)))).))))).))).).)))))...)))))..-............. ( -35.49, z-score = -0.30, R) >droAna3.scaffold_13266 13877511 82 + 19884421 UUCGGAGU-CCAGUCACCAGGA-AUCGAAUUUGGUCUCCAAGCCAUCCGGCCGGGCCAAAGACGAGUGGCAGACUGAGAAAAAA---------- (((..(((-(..(((((.....-.(((..((((((((....(((....))).))))))))..)))))))).))))..)))....---------- ( -27.00, z-score = -1.60, R) >droWil1.scaffold_180745 1099893 73 + 2843958 -----------AUGUGAUGGCAAGUGGAGAGAGUGGACAAACACAUCCACCAUGGCCAAAGGUGGCCAGCAUACUGAAGAGAUA---------- -----------..(((...((..(((((....(((......))).)))))..((((((....)))))))).)))..........---------- ( -21.70, z-score = -1.83, R) >consensus UUCGGAGUGCCGCCCACCUGGAUGUCCGGGCCAAAAUCCAAGCCAUCCGGCCGGGACAAAGGCGAGCGGCAGACUGAGGG_AAAAUCGAGGAAA (((((..((((((.(.(((...(((((.((((................)))).))))).))).).))))))..)))))................ (-19.71 = -23.15 + 3.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:30 2011