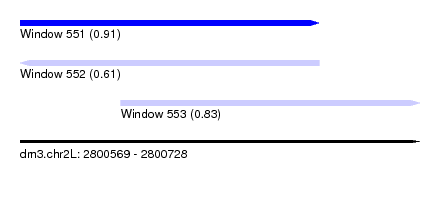
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,800,569 – 2,800,728 |
| Length | 159 |
| Max. P | 0.908620 |

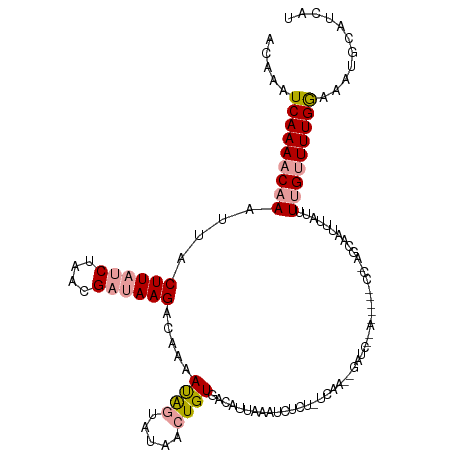
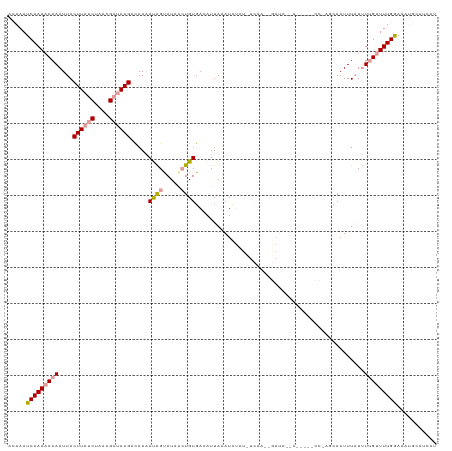
| Location | 2,800,569 – 2,800,688 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 66.48 |
| Shannon entropy | 0.47273 |
| G+C content | 0.29134 |
| Mean single sequence MFE | -20.07 |
| Consensus MFE | -8.99 |
| Energy contribution | -11.33 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2800569 119 + 23011544 AAAAAUCAAAACAAAUUACUUAUCUAACGAUAAGCCAAAAUAGUAUGACUGUGAAAUUAAAUCUUUUUCAAUAGAUCCGAUCCAUCCAGGCAAUUUAUUUGGUUUUGGAAAUGCAUUAU .....(((((((......((((((....))))))...((((((..((.(((.((.(((..((((........))))..)))...))))).))..)))))).)))))))........... ( -19.90, z-score = -1.39, R) >droSec1.super_5 959824 96 + 5866729 ACAAAUCAAAACAAAUUACUUAUCUAACGAUAAGACAAAAUAGUAUAGCUGUGACAUUAAAUCAC-----------------------AGCAAUUUAUUUUGUUUUGGAAAUGCAUGAU .....((((((((((.((((((((....)))))).............(((((((.......))))-----------------------)))....)).))))))))))........... ( -26.30, z-score = -5.69, R) >droEre2.scaffold_4929 2847084 113 + 26641161 GCAAAUCAAAACAAAUUACUUACCUAAAGUGAAGUUAACACGGCAUUAAUGUGACAUUAUAUUUCUCUCAAAGGAUCUAA-----CCGCUCAAUUU-UCUUGCUUUGAAUGCGCAUCAU (((..(((((.(((.((((((.....)))))).........(((..(((((...))))).............((......-----)))))......-..))).))))).)))....... ( -14.00, z-score = 0.42, R) >consensus ACAAAUCAAAACAAAUUACUUAUCUAACGAUAAGACAAAAUAGUAUAACUGUGACAUUAAAUCUCU_UCAA__GAUC__A_____CC_AGCAAUUUAUUUUGUUUUGGAAAUGCAUCAU .....(((((((((....((((((....)))))).....((((.....))))...............................................)))))))))........... ( -8.99 = -11.33 + 2.34)

| Location | 2,800,569 – 2,800,688 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 66.48 |
| Shannon entropy | 0.47273 |
| G+C content | 0.29134 |
| Mean single sequence MFE | -24.13 |
| Consensus MFE | -8.48 |
| Energy contribution | -9.93 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.610657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2800569 119 - 23011544 AUAAUGCAUUUCCAAAACCAAAUAAAUUGCCUGGAUGGAUCGGAUCUAUUGAAAAAGAUUUAAUUUCACAGUCAUACUAUUUUGGCUUAUCGUUAGAUAAGUAAUUUGUUUUGAUUUUU ............((((((((((.....((.(((...((((..(((((........)))))..))))..))).))......))))(((((((....))))))).....))))))...... ( -23.70, z-score = -1.49, R) >droSec1.super_5 959824 96 - 5866729 AUCAUGCAUUUCCAAAACAAAAUAAAUUGCU-----------------------GUGAUUUAAUGUCACAGCUAUACUAUUUUGUCUUAUCGUUAGAUAAGUAAUUUGUUUUGAUUUGU ............((((.((((((((((((((-----------------------(((((.....)))))))).............((((((....)))))).))))))))))).)))). ( -27.70, z-score = -5.41, R) >droEre2.scaffold_4929 2847084 113 - 26641161 AUGAUGCGCAUUCAAAGCAAGA-AAAUUGAGCGG-----UUAGAUCCUUUGAGAGAAAUAUAAUGUCACAUUAAUGCCGUGUUAACUUCACUUUAGGUAAGUAAUUUGUUUUGAUUUGC .......(((.(((((((((((-(......((((-----(.......(((....)))...(((((...)))))..)))))......)))((((.....))))...)))))))))..))) ( -21.00, z-score = 0.14, R) >consensus AUAAUGCAUUUCCAAAACAAAAUAAAUUGCC_GG_____U__GAUC__UUGA_AGAGAUUUAAUGUCACAGUAAUACUAUUUUGACUUAUCGUUAGAUAAGUAAUUUGUUUUGAUUUGU ............(((((((((...................................(((.....))).................(((((((....)))))))..)))))))))...... ( -8.48 = -9.93 + 1.45)

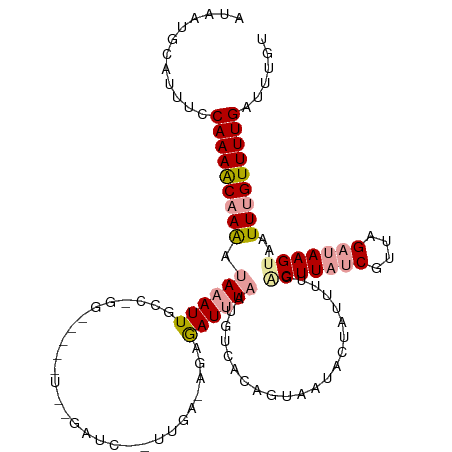
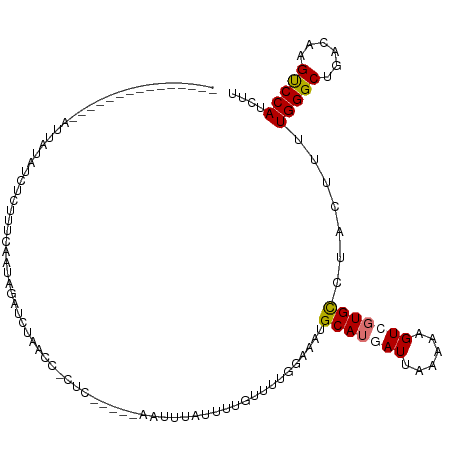
| Location | 2,800,609 – 2,800,728 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 66.42 |
| Shannon entropy | 0.54010 |
| G+C content | 0.35715 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -11.59 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2800609 119 + 23011544 UAGUAUGACUGUGAAAUUAAAUCUUUUUCAAUAGAUCCGAUCCAUCCAGGCAAUUUAUUUGGUUUUGGAAAUGCAUUAUUAAAAGGUCGUGCCUACUUUUGGGCUGACAAGUCCAUCUU ..((((((((.(((((.........)))))..........((((.(((((.......)))))...))))...............)))))))).......((((((....)))))).... ( -26.10, z-score = -0.80, R) >droSim1.chr2L 2758276 93 + 22036055 ---------------------UCUCUUUCAAUAGAUCUAUCCACGC-----AAUUUAUUUUGUUUUGGAAAUGCAUGAUUAAAAAGUCGUGUCUACUUUUGGGCUGACAUGUCCAUCUU ---------------------..........((((....((((.((-----((......))))..))))....(((((((....)))))))))))....(((((......))))).... ( -19.00, z-score = -1.66, R) >droSec1.super_5 959864 96 + 5866729 --------------UAGUAUAGCUGUGACAUUAAAUCACAGC---------AAUUUAUUUUGUUUUGGAAAUGCAUGAUUAAAAAGUCGUGUCUACUUUUGGGCUGACAAGUCCAUCUU --------------.((((..(((((((.......)))))))---------.....................((((((((....)))))))).))))..((((((....)))))).... ( -25.50, z-score = -2.71, R) >droEre2.scaffold_4929 2847124 113 + 26641161 CGGCAUUAAUGUGACAUUAUAUUUCUCUCAAAGGAUCUAACCGCUC-----AAUUU-UCUUGCUUUGAAUGCGCAUCAUGCAAAAGUAUUGCCUACUUUUGGGCUGACAUGCCCAACUU ..((((..(((((.((((..............((......))((..-----.....-....))....))))))))).))))(((((((.....)))))))((((......))))..... ( -24.10, z-score = -0.93, R) >consensus _______________AUUAUAUCUCUUUCAAUAGAUCUAACC_CUC_____AAUUUAUUUUGUUUUGGAAAUGCAUGAUUAAAAAGUCGUGCCUACUUUUGGGCUGACAAGUCCAUCUU ........................................................................(((((((......))))))).......(((((......))))).... (-11.59 = -11.90 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:20 2011