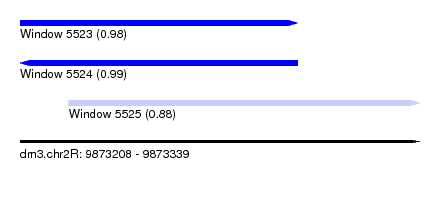
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,873,208 – 9,873,339 |
| Length | 131 |
| Max. P | 0.989270 |

| Location | 9,873,208 – 9,873,299 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.34 |
| Shannon entropy | 0.51261 |
| G+C content | 0.63524 |
| Mean single sequence MFE | -40.63 |
| Consensus MFE | -23.15 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
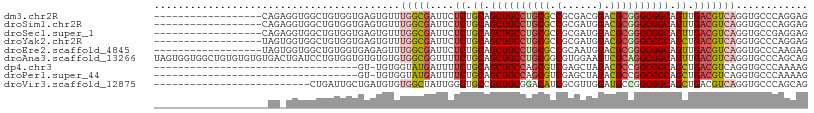
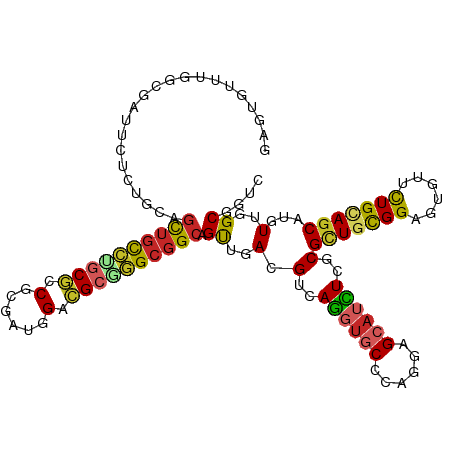
>dm3.chr2R 9873208 91 + 21146708 ------------------CAGAGGUGGCUGUGGUGAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGACGGACGCGGGCGGCAGUUGACGUCAGGUGCCCAGGAG ------------------...(((..((((..(.((((........)))).)..))))..)))((.(((((.....)))))))((((.(((....))).))))...... ( -43.30, z-score = -1.54, R) >droSim1.chr2R 8309822 91 + 19596830 ------------------CAGAGGUGGCUGUGGUGAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGAUGGACGCGGGCGGCAGUUGACGUCAGGUGCCCAGGAG ------------------...(((..((((..(.((((........)))).)..))))..)))((.(((((.....)))))))((((.(((....))).))))...... ( -43.30, z-score = -1.86, R) >droSec1.super_1 7398801 91 + 14215200 ------------------CAGAGGUGGCUGUGGUGAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGAUGGACGCGGGCGGCAGUUGACGUCAGGUGCCGAGGAG ------------------...(((..((((..(.((((........)))).)..))))..)))((.(((((.....)))))))((((.(((....))).))))...... ( -44.20, z-score = -2.40, R) >droYak2.chr2R 14059335 91 - 21139217 ------------------UAGUGGUGGCUGUGGUGAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGAUGGACGCGGGCGGCAGCUGACGUCAGGUGCCCAGGAG ------------------....((..((((..(.((((........)))).)..))))..)).((.(((((.....)))))))((((.(((....))).))))...... ( -43.90, z-score = -1.72, R) >droEre2.scaffold_4845 6641202 91 - 22589142 ------------------UAGUGGUGGCUGUGGUGAGAGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCAAUGGACGCGGGCGGCAGUUGACGUCAGGUGCCCAAGAG ------------------.......((((((...(((((((....))))))).))))))(((((((((.....)).)))))))((((.(((....))).))))...... ( -40.20, z-score = -1.49, R) >droAna3.scaffold_13266 17184714 109 + 19884421 UAGUGGUGGCUGUGUGUGUGACUGAUCCUGUGGUGUGUGUGUGGCGGUUUUCUGCAGCUGCCUGCGGCGUGGAAGUCGCAGGCGGCAGUUGACGUCAGGUGCCCAGCAG ........((((.(..(.((((.....................((((....)))).((((((((((((......)))))))))))).......)))).)..).)))).. ( -47.10, z-score = -1.52, R) >dp4.chr3 7652557 74 - 19779522 ----------------------------------GU-UGUGGUAUGAUUUUCUGCAGCUGCCAGCGUCGAGCUAGACGCCGGCGGCAGCUGACGUCAGGUGCCCAAAAG ----------------------------------.(-((.((((((((..((.((.((((((.(((((......))))).)))))).)).)).))))..)))))))... ( -32.90, z-score = -2.68, R) >droPer1.super_44 304038 74 - 614636 ----------------------------------GU-UGUGGUAUGAUUUUCUGCAGCUGCCAGCGUCGAGCUAGACGCCGGCGGCAGCUGACGUCAGGUGCCCAAAAG ----------------------------------.(-((.((((((((..((.((.((((((.(((((......))))).)))))).)).)).))))..)))))))... ( -32.90, z-score = -2.68, R) >droVir3.scaffold_12875 14500503 83 - 20611582 --------------------------CUGAUUGCUGAUGUGUGGCUAUUGGCUGCCGCUGCGGACAUCGCGUUGGAUGCCGGCGGCAGCUGACGUCAGGUGCCCAGCAG --------------------------.....(((((...(.((((..(..((((((((..(((.((((......)))))))))))))))..).)))).)....))))). ( -37.90, z-score = -1.39, R) >consensus ___________________AG_GGUGGCUGUGGUGAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGAUGGACGCGGGCGGCAGUUGACGUCAGGUGCCCAGGAG .........................................(((((....((.((.((((((((((.(......).)))))))))).)).)))))))............ (-23.15 = -23.27 + 0.11)
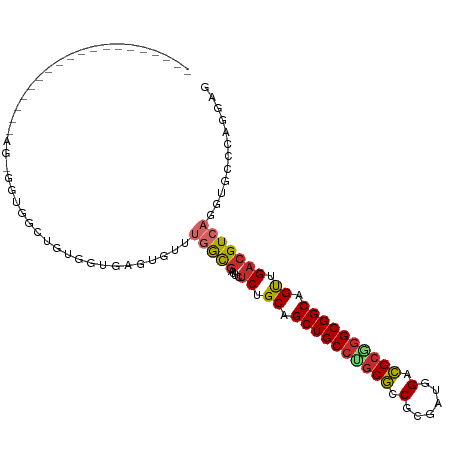
| Location | 9,873,208 – 9,873,299 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 72.34 |
| Shannon entropy | 0.51261 |
| G+C content | 0.63524 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -21.28 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9873208 91 - 21146708 CUCCUGGGCACCUGACGUCAACUGCCGCCCGCGUCCGUCGCGGCGCAGGCAGCUGCAGAGAAUCGCCAAACACUCACCACAGCCACCUCUG------------------ ......(((.......(.((.((((((((((((.....))))).)).))))).))).(((............)))......))).......------------------ ( -28.90, z-score = -0.39, R) >droSim1.chr2R 8309822 91 - 19596830 CUCCUGGGCACCUGACGUCAACUGCCGCCCGCGUCCAUCGCGGCGCAGGCAGCUGCAGAGAAUCGCCAAACACUCACCACAGCCACCUCUG------------------ ......(((.......(.((.((((((((((((.....))))).)).))))).))).(((............)))......))).......------------------ ( -28.90, z-score = -0.94, R) >droSec1.super_1 7398801 91 - 14215200 CUCCUCGGCACCUGACGUCAACUGCCGCCCGCGUCCAUCGCGGCGCAGGCAGCUGCAGAGAAUCGCCAAACACUCACCACAGCCACCUCUG------------------ ......(((.......(.((.((((((((((((.....))))).)).))))).))).(((............)))......))).......------------------ ( -29.20, z-score = -1.77, R) >droYak2.chr2R 14059335 91 + 21139217 CUCCUGGGCACCUGACGUCAGCUGCCGCCCGCGUCCAUCGCGGCGCAGGCAGCUGCAGAGAAUCGCCAAACACUCACCACAGCCACCACUA------------------ ......(((.......(.(((((((((((((((.....))))).)).))))))))).(((............)))......))).......------------------ ( -35.50, z-score = -2.66, R) >droEre2.scaffold_4845 6641202 91 + 22589142 CUCUUGGGCACCUGACGUCAACUGCCGCCCGCGUCCAUUGCGGCGCAGGCAGCUGCAGAGAAUCGCCAAACUCUCACCACAGCCACCACUA------------------ ......(((.......(.((.((((((((((((.....))))).)).))))).))).((((..........))))......))).......------------------ ( -29.50, z-score = -1.18, R) >droAna3.scaffold_13266 17184714 109 - 19884421 CUGCUGGGCACCUGACGUCAACUGCCGCCUGCGACUUCCACGCCGCAGGCAGCUGCAGAAAACCGCCACACACACACCACAGGAUCAGUCACACACACAGCCACCACUA ..((((......((((.....((((.(((((((.(......).)))))))....))))....((.................))....))))......))))........ ( -28.33, z-score = -0.36, R) >dp4.chr3 7652557 74 + 19779522 CUUUUGGGCACCUGACGUCAGCUGCCGCCGGCGUCUAGCUCGACGCUGGCAGCUGCAGAAAAUCAUACCACA-AC---------------------------------- .(((((((((.(((....))).))))(((((((((......))))))))).....)))))............-..---------------------------------- ( -30.90, z-score = -3.32, R) >droPer1.super_44 304038 74 + 614636 CUUUUGGGCACCUGACGUCAGCUGCCGCCGGCGUCUAGCUCGACGCUGGCAGCUGCAGAAAAUCAUACCACA-AC---------------------------------- .(((((((((.(((....))).))))(((((((((......))))))))).....)))))............-..---------------------------------- ( -30.90, z-score = -3.32, R) >droVir3.scaffold_12875 14500503 83 + 20611582 CUGCUGGGCACCUGACGUCAGCUGCCGCCGGCAUCCAACGCGAUGUCCGCAGCGGCAGCCAAUAGCCACACAUCAGCAAUCAG-------------------------- .((((((((...((....))(((((((((((((((......)))).)))..)))))))).....)))......))))).....-------------------------- ( -33.40, z-score = -2.37, R) >consensus CUCCUGGGCACCUGACGUCAACUGCCGCCCGCGUCCAUCGCGGCGCAGGCAGCUGCAGAGAAUCGCCAAACACUCACCACAGCCACC_CU___________________ .(((((((((..((....))..))))(((.(((((......))))).))).....))).))................................................ (-21.28 = -20.60 + -0.68)
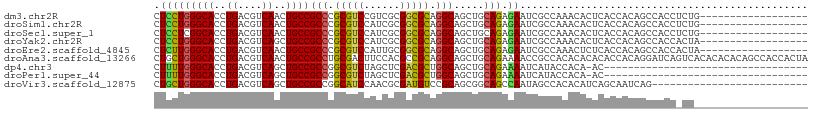
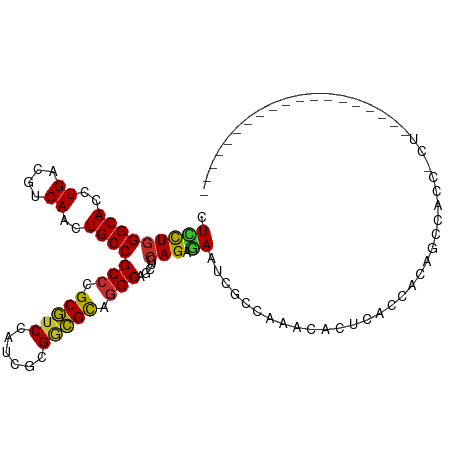
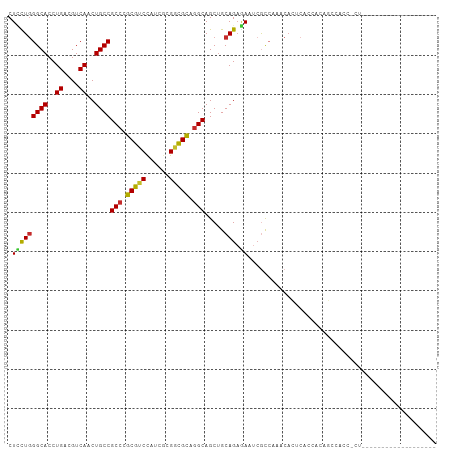
| Location | 9,873,224 – 9,873,339 |
|---|---|
| Length | 115 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.23 |
| Shannon entropy | 0.49368 |
| G+C content | 0.61667 |
| Mean single sequence MFE | -49.33 |
| Consensus MFE | -35.12 |
| Energy contribution | -35.88 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.884774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
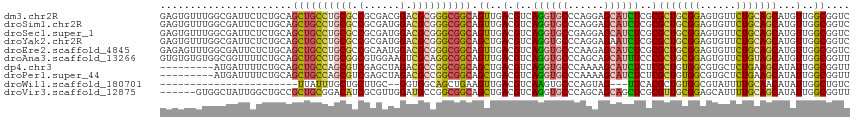
>dm3.chr2R 9873224 115 + 21146708 GAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGACGGACGCGGGCGGCAGUUGACGUCAGGUGCCCAGGAGCAUCUCGCGCUGCGGAGUGUUCUGCAGCAUGUUGGCGGUC (((.(((....))).)))....(((((((((((((...))).)))))))))).((..((((.((((((......)))))).))(((((((......)))))))..))..)).... ( -54.90, z-score = -0.91, R) >droSim1.chr2R 8309838 115 + 19596830 GAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGAUGGACGCGGGCGGCAGUUGACGUCAGGUGCCCAGGAGCAUCUCGCGCUGCGGAGUGUUCUGCAGCAUGUUGGCGGUC (((.(((....))).)))....((((((((((((.....)).)))))))))).((..((((.((((((......)))))).))(((((((......)))))))..))..)).... ( -53.70, z-score = -0.77, R) >droSec1.super_1 7398817 115 + 14215200 GAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGAUGGACGCGGGCGGCAGUUGACGUCAGGUGCCGAGGAGCAUCUCGCGCUGCGGAGUGUUCUGCAGCAUGUUGGCGGUC (((.(((....))).)))....((((((((((((.....)).)))))))))).((..((((.((((((......)))))).))(((((((......)))))))..))..)).... ( -53.70, z-score = -0.76, R) >droYak2.chr2R 14059351 115 - 21139217 GAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGAUGGACGCGGGCGGCAGCUGACGUCAGGUGCCCAGGAGAAUCUCGCGCUGCGGAGUGUUCUGCAGCAUGUUGGCGGUC ((.((.(..((((((((((.((((((((.((.(((((.....)))))))))))))))......((...)).))))))))....(((((((......)))))))..))..))).)) ( -57.20, z-score = -1.79, R) >droEre2.scaffold_4845 6641218 115 - 22589142 GAGAGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCAAUGGACGCGGGCGGCAGUUGACGUCAGGUGCCCAAGAGCAUCUCGCGCUGCGGAGUGUUCUGCAGCAUGUUGGCGGUC (((((((....)))))))....((((((((((((.....)).)))))))))).((..((((.((((((......)))))).))(((((((......)))))))..))..)).... ( -57.80, z-score = -2.45, R) >droAna3.scaffold_13266 17184748 115 + 19884421 GUGUGUGUGGCGGUUUUCUGCAGCUGCCUGCGGCGUGGAAGUCGCAGGCGGCAGUUGACGUCAGGUGCCCAGCAGCAUUUCCCGCUGCGGAGUGUUCUGUAGCAUGUUGGCGGUU .........((((....)))).((((((((((((......)))))))))))).((..(((..((((((......))))))..)(((((((......)))))))..))..)).... ( -53.30, z-score = -1.97, R) >dp4.chr3 7652565 106 - 19779522 ---------AUGAUUUUCUGCAGCUGCCAGCGUCGAGCUAGACGCCGGCGGCAGCUGACGUCAGGUGCCCAAAAGCAUCUCUCGCUGUGGCGUGCUCUGAAGCAUAUUGGCGGUU ---------........((((.((((((.(((((......))))).)))))).(((....(((((..((((..(((.......))).))).)..).)))))))......)))).. ( -43.60, z-score = -0.98, R) >droPer1.super_44 304046 106 - 614636 ---------AUGAUUUUCUGCAGCUGCCAGCGUCGAGCUAGACGCCGGCGGCAGCUGACGUCAGGUGCCCAAAAGCAUCUCUCGCUGUGGCGUGCUCUGAAGCAUAUUGGCGGUU ---------........((((.((((((.(((((......))))).)))))).(((....(((((..((((..(((.......))).))).)..).)))))))......)))).. ( -43.60, z-score = -0.98, R) >droWil1.scaffold_180701 1092263 87 + 3904529 -----------------------UUAUUUGCUGCUUGC--GGUGGCAGCUGAAGUUGACGUCAAGUGCCCAGUAG---UUCACGCUGUGGCGUAUUUUGCAACAUAUUGGCUGUC -----------------------......((((....)--)))((((((..(.((((.((..(..((((((((..---.....)))).))))..)..))))))...)..)))))) ( -26.80, z-score = -0.20, R) >droVir3.scaffold_12875 14500517 109 - 20611582 ------GUGGCUAUUGGCUGCCGCUGCGGACAUCGCGUUGGAUGCCGGCGGCAGCUGACGUCAGGUGCCCAGCAGCAGCUCGCGUUGCGGAGCAUUUUGCAGCAUAUUGGCGGUU ------...((((...(((((.(((((((.((((((((..(.((((...)))).)..))))..)))).)).))))).((((((...)).)))).....)))))....)))).... ( -48.70, z-score = -0.22, R) >consensus GAGUGUUUGGCGAUUCUCUGCAGCUGCCUGCGCCGCGAUGGACGCGGGCGGCAGUUGACGUCAGGUGCCCAGGAGCAUCUCGCGCUGCGGAGUGUUCUGCAGCAUGUUGGCGGUC ................((.((.((((((((((.(......).)))))))))).)).))((((((((((......)))))....(((((((......)))))))....)))))... (-35.12 = -35.88 + 0.76)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:28 2011