| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,869,938 – 9,870,047 |
| Length | 109 |
| Max. P | 0.884337 |

| Location | 9,869,938 – 9,870,047 |
|---|---|
| Length | 109 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.43 |
| Shannon entropy | 0.73729 |
| G+C content | 0.55798 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -14.01 |
| Energy contribution | -13.91 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 2.10 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.884337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
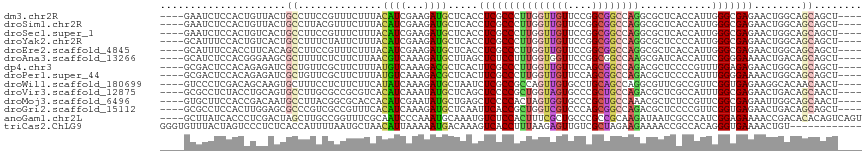
>dm3.chr2R 9869938 109 + 21146708 ----GAAUCUCCACUGUUACUGCCUUCCGUUUCUUUACAUCGAAGAUGCUCACCUCGCCCUUGGUUGUUCCGGCGGCCAGGCGCUCACCAUUGGGCGAGAACUGGCAGCAGCU---- ----...........(((.(((((....((.(((((.....))))).))....(((((((.(((((((....)))))))((......))...)))))))....))))).))).---- ( -38.00, z-score = -1.83, R) >droSim1.chr2R 8306547 109 + 19596830 ----GAAUCUCCACUGUUACUGCCUUACGUUUCUUUACAUCGAAGAUGCUCACCUCGCCCUUGGUUGUUCCGGCGGCCAGGCGCUCACCAUUGGGCGAGAACUGGCAGCAGCU---- ----...........(((.(((((....((.(((((.....))))).))....(((((((.(((((((....)))))))((......))...)))))))....))))).))).---- ( -38.00, z-score = -1.99, R) >droSec1.super_1 7395530 109 + 14215200 ----GAAUCUCCACUGUCACUGCCUUCCGUUUCUUUACAUCGAAGAUGCUCACCUCGCCCUUGGUUGUUCCGGCGGCCAGGCGCUCACCAUUGGGCGAGAACUGGCAGCAGCU---- ----...........((..(((((....((.(((((.....))))).))....(((((((.(((((((....)))))))((......))...)))))))....)))))..)).---- ( -37.10, z-score = -1.38, R) >droYak2.chr2R 14056051 109 - 21139217 ----GCAUUUCCACUGUCACUGCCUUUCUAUUCUUUACAUCGAAGAUGCUCACCUCGCCCUUGGUUGUUCCGGCGGCCAGGCGCUCCCCAUUGGGCGAGAACUGGCAGCAGCU---- ----...........((..(((((.......(((((.....))))).......(((((((.(((((((....)))))))((......))...)))))))....)))))..)).---- ( -34.70, z-score = -0.94, R) >droEre2.scaffold_4845 6637925 109 - 22589142 ----GCAUUUCCACCUUCACAGCCUUCCGUUUCUUUACAUCGAAGAUGCUCACCUCGCCCUUGGUUGUUCCGGCGGCCAGGCGCUCACCAUUGGGCGAGAACUGGCAGCAGCU---- ----.................(((....((.(((((.....))))).))....(((((((.(((((((....)))))))((......))...)))))))....))).......---- ( -32.50, z-score = -0.40, R) >droAna3.scaffold_13266 17181450 109 + 19884421 ----GCAUCUCCACGGGAAGCGCUUUUCUCUUCUUAACGUCAAAGAUGCUUAGCUCUCCUUUGGUGGUUCCGGCGGCCAAGCGAUCACCAUUCGGGGAAAACUGACAGCAGCU---- ----((((((..((((((((.(.....).)))))...)))...)))))).....(((((..((((((((...((......))))))))))...)))))...(((....)))..---- ( -30.50, z-score = 0.08, R) >dp4.chr3 7649482 109 - 19779522 ----GCGACUCCACAGAGAUCGCUGUUCGCUUCUUUAUGUCAAAGACGCUCACUUCGCCCUUGGUUGUUCCAGCGGCCAGACGCUCCCCGUUUGGAGAGAACUGGCAGCAGCU---- ----..(((((....))).))((((((.((.(((((.....))))).))......(((...(((.....))))))(((((...(((.((....)).)))..))))))))))).---- ( -33.80, z-score = -0.22, R) >droPer1.super_44 300963 109 - 614636 ----GCGACUCCACAGAGAUCGCUGUUCGCUUCUUUAUGUCAAAGACGCUCACUUCGCCCUUGGUUGUUCCAGCGGCCAGACGCUCCCCAUUUGGGGAAAACUGGCAGCAGCU---- ----..(((((....))).))((((((.((.(((((.....))))).)).......(((.((((((((....))))))))....(((((....))))).....))))))))).---- ( -37.10, z-score = -1.47, R) >droWil1.scaffold_180699 1196008 109 + 2593675 ----GUCCCUCGACAGCAAGUGCUUUCCUCUUCUUCAUAUCAAAGAUGCUAAUCUCGCCGCCAGUUGUGCCUGCAGCCAGGCGUUCGCCGUUCGGUGAGAAGGCACAACAACU---- ----(((....))).....((((((((................((((....))))...((((.(((((....)))))..)))).(((((....))))))))))))).......---- ( -31.70, z-score = -1.22, R) >droVir3.scaffold_12875 14496989 109 - 20611582 ----GCGCCUCUACCUGCAGUGCCUUGCGCCGCGUCACAUCAAAUAUGCUCAGCUCCCCGCUGGUAGUGCCCGCUGCCAGACGCUCGCCAUUUGGCGAGAACUGACAGCAACU---- ----((((........((.((((...)))).)).............(((.((((.....)))))))))))..((((.(((...((((((....))))))..))).))))....---- ( -39.80, z-score = -2.06, R) >droMoj3.scaffold_6496 23926057 109 + 26866924 ----GUGCUUCCACCGACAAUGCCUUACGGCGCACCACAUCGAAUAUGCUGAGCUCCCCACUAGUGGUGCCCGCUGCCAAACGCUCUCCGUUCGGCGAGAAUUGGCAGCAACU---- ----(((....))).......(((....)))(((((((.........((...)).........)))))))..((((((((...(((.((....)).)))..))))))))....---- ( -37.67, z-score = -2.43, R) >droGri2.scaffold_15112 4157101 109 - 5172618 ----GCGCCUCCACUUGGAGCGCCCGUCGCCGUUUCACAUCAAAGAUGCUCAAUUCACCGCUGGUCGUCCCAGCGGCCAGACGCUCCCCGUUCGGUGAGAACUGACAGCAGCU---- ----((((.(((....))))))).....((.((((((((((...)))).....(((((((((((((((....))))))))(((.....))).)))))))...))).))).)).---- ( -37.20, z-score = -1.27, R) >anoGam1.chr2L 2802251 113 - 48795086 ----GCUUAUCACCCUCGACUAGCUUGCCGGUUUCGCAAUCCCAAAUGCAAAUGUCUCCACUUUCGCUGCCCGCCGCAAGAUAAUCGCCCAUCGGAGAAAACCGACACACAGUCAGU ----.............((((.(..((.((((((.(((........))).....(((((......((.(....).))..(((........)))))))))))))).))..)))))... ( -24.30, z-score = -0.96, R) >triCas2.ChLG9 6472714 105 + 15222296 GGGUGUUUACUAGUCCCUCUCACCAUUUUAAUGCUAACAUUAAAAAUGACAAAGUCACCUUUAAGAGUUGUCGCUAGAAGAAAACCGCCACAGGGUGAAAACUGU------------ .(((((((.(((((..(.(((....((((((((....)))))))).((((...)))).......)))..)..))))).)))....))))((((........))))------------ ( -21.80, z-score = -0.57, R) >consensus ____GCAUCUCCACCGUCACUGCCUUUCGCUUCUUUACAUCAAAGAUGCUCACCUCGCCCUUGGUUGUUCCGGCGGCCAGACGCUCACCAUUGGGCGAGAACUGGCAGCAGCU____ .....................((..............((((...)))).....(((((((((((((((....))))))))............)))))))........))........ (-14.01 = -13.91 + -0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:26 2011