| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,861,036 – 9,861,126 |
| Length | 90 |
| Max. P | 0.999820 |

| Location | 9,861,036 – 9,861,126 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 72.07 |
| Shannon entropy | 0.55684 |
| G+C content | 0.49888 |
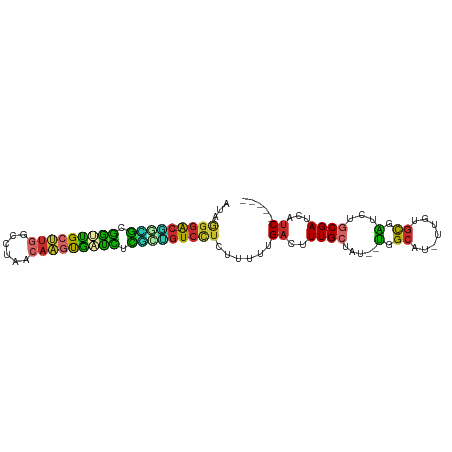
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -22.08 |
| Energy contribution | -20.48 |
| Covariance contribution | -1.60 |
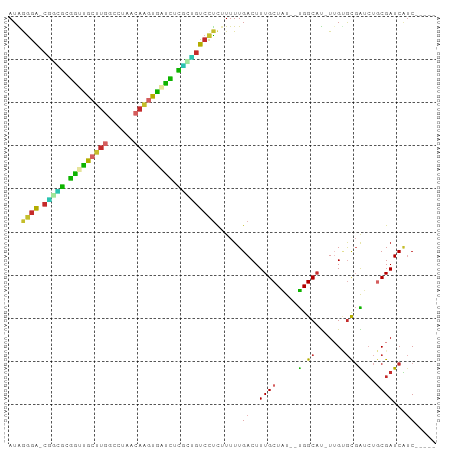
| Combinations/Pair | 1.93 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.48 |
| SVM RNA-class probability | 0.999820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
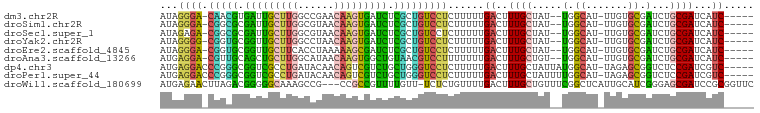
>dm3.chr2R 9861036 90 - 21146708 AUAGGGA-CAACGUGAUUGCUUGGCCGAACAAGUGAUCUCGCUGUCCUCUUUUUGACUUUGCUAU--UGGCAU-UUGUGCGAUCUGCGAUCAUC----- ...((((-((.((.(((..((((......))))..))).)).)))))).....(((..((((...--(.(((.-...))).)...)))))))..----- ( -30.40, z-score = -3.18, R) >droSim1.chr2R 8297202 90 - 19596830 AUAGGGA-CGGCGCGAUUGCUUGGCGUAACAAGUGAUCUCGCUGUCCUCUUUUUGACUUUGCUAU--UGGCAU-UUGUGCGAUCUGCGAUCAUC----- ...((((-(((((.(((..((((......))))..))).))))))))).....(((..((((...--(.(((.-...))).)...)))))))..----- ( -36.00, z-score = -4.28, R) >droSec1.super_1 7386674 90 - 14215200 AUAGAGA-CGGCGCGAUUGCUUGGCGUAACAAGUGAUCUCGCUGUCCUCUUUUUGACUUUGCUAU--UGGCAU-UUGUGCGAUCUGCGAUCAUC----- ..(((((-(((((.(((..((((......))))..))).)))))).))))...(((..((((...--(.(((.-...))).)...)))))))..----- ( -33.30, z-score = -3.36, R) >droYak2.chr2R 14046654 90 + 21139217 AUAGGGG-CGGUGCGGUUGCUUGGCCUAACAAGUGAUCUCGCUGUCCUCUUUUUGACUUUGCUAU--UGGCAU-UUGUGCGAUCUGCGAUCAUC----- ...((((-(((((.(((..((((......))))..))).))))))))).....(((..((((...--(.(((.-...))).)...)))))))..----- ( -32.90, z-score = -3.11, R) >droEre2.scaffold_4845 6628664 90 + 22589142 AUAGGGA-CGGUGCGGUUGCUUCACCUAAAAAGCGAUCUCGCUGUCCUCUUUUUGACUUUGCUAU--UGGCAU-UUGUGCGAUCUGCGAUCAUC----- ...((((-(((((.((((((((........)))))))).))))))))).....(((..((((...--(.(((.-...))).)...)))))))..----- ( -31.20, z-score = -3.40, R) >droAna3.scaffold_13266 17172685 90 - 19884421 AUGAGGA-CGUUGCAGCUGCUUGGCAUAACAAGUGGCUGUAACGUCCUUUUUUUGACUUUGCUGU--UGGCAU-UUGUGCGAUCUGCGAUCAUC----- ..(((((-(((((((((..((((......))))..))))))))))))))....(((..((((...--(.(((.-...))).)...)))))))..----- ( -41.50, z-score = -6.18, R) >dp4.chr3 7641597 93 + 19779522 AUGAGGACCCGGGCGGUCGCCUGAUACAACAGUCGUCUGCUGGGUCCUCUUUUUGACUUUGCUAUUAUGGCAU-UAGAGCGGUCUCCGAUCGUC----- ..((((((((((.(((.((.(((......))).)).))))))))))))).......((.((((.....)))).-.)).((((((...)))))).----- ( -37.30, z-score = -3.08, R) >droPer1.super_44 293112 93 + 614636 AUGAGGACCCGGGCGGUCGCCUGAUACAACAGUCGUCUGCUGGGUCCUCUUUUUGACUUUGCUAUUUUGGCAU-UAGAGCGGUCUCCGAUCGUC----- ..((((((((((.(((.((.(((......))).)).))))))))))))).......((.((((.....)))).-.)).((((((...)))))).----- ( -37.30, z-score = -3.17, R) >droWil1.scaffold_180699 1186891 95 - 2593675 AUGAGAACUUAGACGGGGGCAAAGCCG---CCGCCGUUUUGUU-UCUCUGUUUUGACUUUGCUGUUUCGGCUCAUUGCAUCGGGAGCGAUCCGCGGUUC ..(((((...((((((.(((......)---)).))))))...)-)))).((..(((....((((...)))))))..))(((((((....))).)))).. ( -31.50, z-score = -0.70, R) >consensus AUAGGGA_CGGCGCGGUUGCUUGGCCUAACAAGUGAUCUCGCUGUCCUCUUUUUGACUUUGCUAU__UGGCAU_UUGUGCGAUCUGCGAUCAUC_____ ...((((.(((((.(((((((((......))))))))).)))))))))...........(((((...)))))........((((...))))........ (-22.08 = -20.48 + -1.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:25 2011