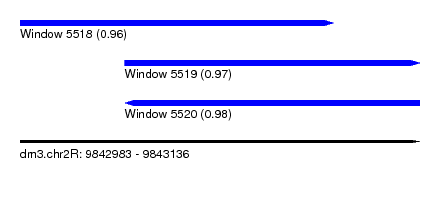
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,842,983 – 9,843,136 |
| Length | 153 |
| Max. P | 0.984129 |

| Location | 9,842,983 – 9,843,103 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.78 |
| Shannon entropy | 0.36419 |
| G+C content | 0.49057 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -20.64 |
| Energy contribution | -20.44 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
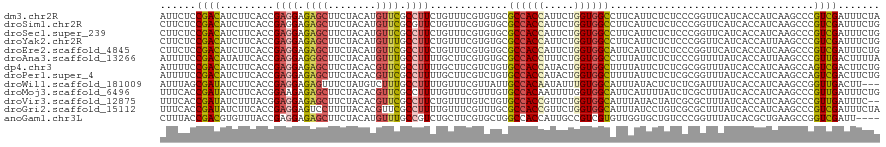
>dm3.chr2R 9842983 120 + 21146708 AUUCUCCGACAUCUUCACCGAGGAGAGCUUCUACAUGUUCGCCUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCCUUCAUUCUCUCCCGGUUCAUCACCAUCAAGCCCGUCGAUUUCUA ......((((......((((.((((((....((((((...((.....))..)))))).((((((.....)))))).......)))))))))).......(.....)...))))....... ( -33.10, z-score = -2.96, R) >droSim1.chr2R 8277495 120 + 19596830 CUUCUCCGACAUCUUCACCGAGGAGAGCUUCUACAUGUUCGCGUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCCUUCAUUCUCUCCCGGUUCAUCACCAUCAAGCCCGUCGAUUUCUG ......((((......((((.((((((.........((.((((........)))).))((((((.....)))))).......)))))))))).......(.....)...))))....... ( -34.80, z-score = -3.04, R) >droSec1.super_239 13203 120 + 19797 CUUCUCCGACAUCUUCACCGAGGAGAGCUUCUACAUGUUCGCCUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCCUUCAUUCUCUCCCGGUUCAUCACCAUCAAGCCCGUCGAUUUCUG ......((((......((((.((((((....((((((...((.....))..)))))).((((((.....)))))).......)))))))))).......(.....)...))))....... ( -33.10, z-score = -2.86, R) >droYak2.chr2R 14028096 120 - 21139217 CUUCUCCGACAUCUUCACCGAGGAGAGCUUCUACAUGUUUGCCUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCCUUCAUUCUCUCCCGGUUCAUCACCAUUAAGCCCGUCGAUUUCUG ......((((......((((.((((((....((((((...((.....))..)))))).((((((.....)))))).......)))))))))).......(.....)...))))....... ( -33.80, z-score = -3.29, R) >droEre2.scaffold_4845 6609975 120 - 22589142 CUUCUCCGACAUCUUCACCGAGGAGAGCUUCUACAUGUUCGCCUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCAUUCAUUCUCUCCCGGUUCAUCACCAUCAAGCCCGUCGAUUUCUG ......((((......((((.((((((....((((((...((.....))..)))))).((((((.....)))))).......)))))))))).......(.....)...))))....... ( -34.00, z-score = -3.08, R) >droAna3.scaffold_13266 17153224 120 + 19884421 AUUUUCCGACAUAUUCACCGAGGAGGGCUUCUACAUGUUUGCCUUUUGCUUCGUGUGCGCCACCUUUCUGGUGGCCUUUAUUCUCUCCCGUUUUAUCACCAUUAAGCCCGUUGACUUUUA ......((((...(((.....)))((((((.((((((...((.....))..)))))).((((((.....))))))............................))))))))))....... ( -30.80, z-score = -2.92, R) >dp4.chr3 8156227 120 + 19779522 AUUUUCCGACAUCUUCACCGAGGAGAGCUUCUACACGUUCGCCUUUUGCUUCGUCUGUGCCACCAUACUGGUGGCUUUUAUUCUCUCGCGGUUUAUCACCAUCAAGCCAGUCGACUUCUG ......((((.........((((.((((........)))).))))..((((.......((((((.....))))))..............(((.....)))...))))..))))....... ( -29.90, z-score = -2.10, R) >droPer1.super_4 2033727 120 + 7162766 AUUUUCCGACAUCUUCACCGAGGAGAGCUUCUACACGUUCGCCUUUUGCUUCGUCUGUGCCACCAUACUGGUGGCUUUUAUUCUCUCGCGGUUUAUCACCAUCAAGCCAGUCGACUUCUG ......((((.........((((.((((........)))).))))..((((.......((((((.....))))))..............(((.....)))...))))..))))....... ( -29.90, z-score = -2.10, R) >droWil1.scaffold_181009 1337200 117 + 3585778 AUUUAGCGAUAUCUUCACCGAGGAGAGUUUCUAUGUCUUUGCCUUUUGUUUCGUUAUUGCCACAAUAUUUGUGGCAUUUAUACUCUCUCGAUUUAUCACCAUCAAGCCGGUUGACUU--- ...((((((.........((((((.(.......).)))))).........)))))).(((((((.....)))))))...................(((((........)).)))...--- ( -22.37, z-score = -0.76, R) >droMoj3.scaffold_6496 23814317 120 + 26866924 UUUCACCGAUAUCUUCACGGAAGAGAGCUUCUACACGUUCGCCUUUUGUUUCGUUUGUGCCACAAUUUUGGUGGCAUUCAUUUUAUCUCGCUUUAUCACCAUCAAGCCCGUUGAUUUCUG .......((.(((...((((..((((........(((...((.....))..)))..(((((((.......)))))))........))))((((.((....)).)))))))).))).)).. ( -23.80, z-score = -1.21, R) >droVir3.scaffold_12875 14393407 118 - 20611582 UUUCACCGAUAUCUUUACGGAGGAGAGCUUCUACACGUUCGCCUUCUGUUUUGUCUGUGCCACCGUUCUGGUGGCAUUUAUACUAUCGCGCUUUAUCACCAUCAAGCCCGUUGAUUUC-- ..(((.(((((.....(((((((.((((........))))..)))))))..(((..((((((((.....))))))))..))).))))(.((((.((....)).)))).)).)))....-- ( -30.90, z-score = -2.30, R) >droGri2.scaffold_15112 4065401 120 - 5172618 UUUCACCGAUAUCUUCACCGAGGAGUCCUUUUACACGUUCGCCUUUUGUUUCGUUUGCGCCACCGUUCUGGUGGCAUUUAUCCUGUCGCGCUUUAUCACCAUCAAGCCCGUCGAUUUCUA .......((.(((.......((((..........(((...((.....))..)))....((((((.....)))))).....))))(.((.((((.((....)).)))).)).)))).)).. ( -23.50, z-score = -0.60, R) >anoGam1.chr3L 9855921 116 - 41284009 CUUUACCGACGUGUUUACCGAGGAGAGCUUCUACAUGUUUGCCGUCUGCUUCGUGCUGGCCACCAUUGCCGUCGUGUUGGUGCUGUCCCGGUUUAUCACGCUGAAGCCGGUCGAUU---- ......(((((((...((((.(((.(((.((.(((((...((((.......)).)).(((.......)))..))))).)).))).)))))))....)))((....))..))))...---- ( -37.30, z-score = -1.01, R) >consensus AUUCUCCGACAUCUUCACCGAGGAGAGCUUCUACAUGUUCGCCUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCAUUCAUUCUCUCCCGGUUUAUCACCAUCAAGCCCGUCGAUUUCUG ......((((.........((((.((((........)))).)))).............((((((.....))))))..................................))))....... (-20.64 = -20.44 + -0.20)
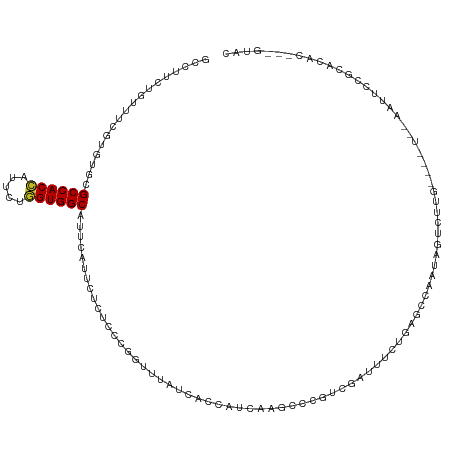
| Location | 9,843,023 – 9,843,136 |
|---|---|
| Length | 113 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.12 |
| Shannon entropy | 0.55235 |
| G+C content | 0.47251 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -11.22 |
| Energy contribution | -11.17 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.965003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
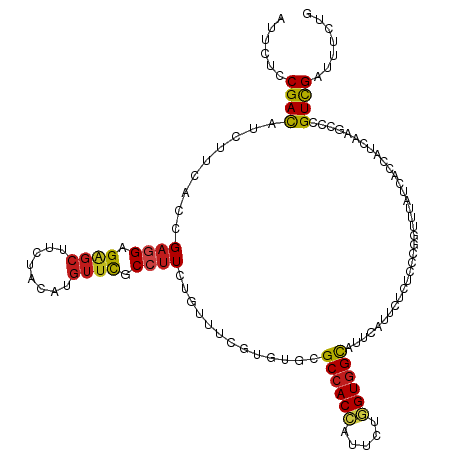
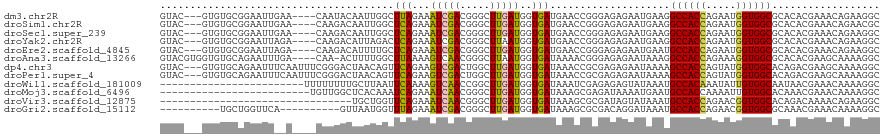
>dm3.chr2R 9843023 113 + 21146708 GCCUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCCUUCAUUCUCUCCCGGUUCAUCACCAUCAAGCCCGUCGAUUUCUAAGCCAAUUGUAUUG----UUCAAUUCCGCACAC---GUAC ...........(((((((((((((.....))))))..............(((.....)))....(((....(((((........)))))....)----)).......))))))---)... ( -26.30, z-score = -2.82, R) >droSim1.chr2R 8277535 113 + 19596830 GCGUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCCUUCAUUCUCUCCCGGUUCAUCACCAUCAAGCCCGUCGAUUUCUGAGCCAAUUGUCUUG----UUCAAUUCCGCACAC---GUAC ...........(((((((((((((.....))))))..............((((((.....(((.........)))...))))))(((((.....----..)))))..))))))---)... ( -31.00, z-score = -3.29, R) >droSec1.super_239 13243 113 + 19797 GCCUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCCUUCAUUCUCUCCCGGUUCAUCACCAUCAAGCCCGUCGAUUUCUGAGCCAAUUGUCUUG----UUCAAUUCCGCACAC---GUAC ...........(((((((((((((.....))))))..............((((((.....(((.........)))...))))))(((((.....----..)))))..))))))---)... ( -31.00, z-score = -3.73, R) >droYak2.chr2R 14028136 113 - 21139217 GCCUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCCUUCAUUCUCUCCCGGUUCAUCACCAUUAAGCCCGUCGAUUUCUGAGUCUAAUGUCUUG----UCUAAUUCCGCACAC---GUAC ...........(((((((((((((.....))))))..............((.....((.(((((..(.((........)).)..)))))...))----.......))))))))---)... ( -28.20, z-score = -3.21, R) >droEre2.scaffold_4845 6610015 113 - 22589142 GCCUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCAUUCAUUCUCUCCCGGUUCAUCACCAUCAAGCCCGUCGAUUUCUGAGCAAAAUGUCUUG----UCUAAUUCCGCACAC---GUAC ...........(((((((((((((.....))))))...............(((((.....(((.........)))...)))))...........----.........))))))---)... ( -28.20, z-score = -2.64, R) >droAna3.scaffold_13266 17153264 115 + 19884421 GCCUUUUGCUUCGUGUGCGCCACCUUUCUGGUGGCCUUUAUUCUCUCCCGUUUUAUCACCAUUAAGCCCGUUGACUUUUAAGCCAAAAGU-UUG----UCAAAUUCUGCACACCACGUAC ......(((...((((((((((((.....))))))..................................(..(((((((.....))))))-)..----.).......))))))...))). ( -26.00, z-score = -2.83, R) >dp4.chr3 8156267 117 + 19779522 GCCUUUUGCUUCGUCUGUGCCACCAUACUGGUGGCUUUUAUUCUCUCGCGGUUUAUCACCAUCAAGCCAGUCGACUUCUGAACUGUUAGUCCCGAAAUUGAAAUUCUGCACAC---GUAC ......(((.(((.(((.((((((.....))))))..............(((.....))).......))).))).....(((...(((((......)))))..))).)))...---.... ( -23.20, z-score = -0.26, R) >droPer1.super_4 2033767 117 + 7162766 GCCUUUUGCUUCGUCUGUGCCACCAUACUGGUGGCUUUUAUUCUCUCGCGGUUUAUCACCAUCAAGCCAGUCGACUUCUGAACUGUUAGUCCCGAAAUUGAAAUUCUGCACAC---GUAC ......(((.(((.(((.((((((.....))))))..............(((.....))).......))).))).....(((...(((((......)))))..))).)))...---.... ( -23.20, z-score = -0.26, R) >droWil1.scaffold_181009 1337240 96 + 3585778 GCCUUUUGUUUCGUUAUUGCCACAAUAUUUGUGGCAUUUAUACUCUCUCGAUUUAUCACCAUCAAGCCGGUUGACUUUUGAAUUAAGCAAAAAAAA------------------------ ...((((((((.((...(((((((.....))))))).....))....((((....(((((........)).)))...))))...))))))))....------------------------ ( -18.90, z-score = -1.98, R) >droMoj3.scaffold_6496 23814357 95 + 26866924 GCCUUUUGUUUCGUUUGUGCCACAAUUUUGGUGGCAUUCAUUUUAUCUCGCUUUAUCACCAUCAAGCCCGUUGAUUUCUGAUUUGUGAGCCAACA------------------------- ............(((.(((((((.......))))))).........(((((...((((..(((((.....)))))...))))..)))))..))).------------------------- ( -20.50, z-score = -1.64, R) >droVir3.scaffold_12875 14393447 88 - 20611582 GCCUUCUGUUUUGUCUGUGCCACCGUUCUGGUGGCAUUUAUACUAUCGCGCUUUAUCACCAUCAAGCCCGUUGAUUUCUGAACCAGCA-------------------------------- ...........(((..((((((((.....))))))))..))).......(((...(((..(((((.....)))))...)))...))).-------------------------------- ( -20.30, z-score = -1.66, R) >droGri2.scaffold_15112 4065441 100 - 5172618 GCCUUUUGUUUCGUUUGCGCCACCGUUCUGGUGGCAUUUAUCCUGUCGCGCUUUAUCACCAUCAAGCCCGUCGAUUUCUAAACCAUUAAC----------UGAACCAGCA---------- ((.(((.(((..(((((.((((((.....))))))........((.((.((((.((....)).)))).)).)).....)))))....)))----------.)))...)).---------- ( -19.90, z-score = -1.04, R) >consensus GCCUUCUGUUUCGUGUGCGCCACCAUUCUGGUGGCAUUCAUUCUCUCCCGGUUUAUCACCAUCAAGCCCGUCGAUUUCUGAGCCAAUAGUCUUG____U__AAUUCCGCACAC___GUAC ..................((((((.....))))))..................................................................................... (-11.22 = -11.17 + -0.05)

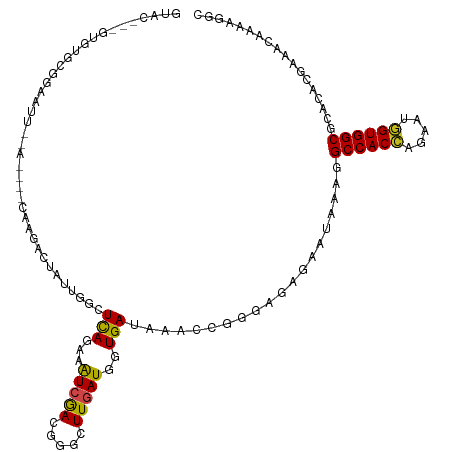
| Location | 9,843,023 – 9,843,136 |
|---|---|
| Length | 113 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.12 |
| Shannon entropy | 0.55235 |
| G+C content | 0.47251 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -14.13 |
| Energy contribution | -13.61 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9843023 113 - 21146708 GUAC---GUGUGCGGAAUUGAA----CAAUACAAUUGGCUUAGAAAUCGACGGGCUUGAUGGUGAUGAACCGGGAGAGAAUGAAGGCCACCAGAAUGGUGGCGCACACGAAACAGAAGGC ...(---((((((..(((((..----.....)))))..............((..(((..((((.....))))...)))..))...((((((.....)))))))))))))........... ( -34.10, z-score = -3.06, R) >droSim1.chr2R 8277535 113 - 19596830 GUAC---GUGUGCGGAAUUGAA----CAAGACAAUUGGCUCAGAAAUCGACGGGCUUGAUGGUGAUGAACCGGGAGAGAAUGAAGGCCACCAGAAUGGUGGCGCACACGAAACAGAACGC ...(---((((((....((((.----(((.....)))..)))).......((..(((..((((.....))))...)))..))...((((((.....)))))))))))))........... ( -36.10, z-score = -3.03, R) >droSec1.super_239 13243 113 - 19797 GUAC---GUGUGCGGAAUUGAA----CAAGACAAUUGGCUCAGAAAUCGACGGGCUUGAUGGUGAUGAACCGGGAGAGAAUGAAGGCCACCAGAAUGGUGGCGCACACGAAACAGAAGGC ...(---((((((....((((.----(((.....)))..)))).......((..(((..((((.....))))...)))..))...((((((.....)))))))))))))........... ( -36.10, z-score = -3.16, R) >droYak2.chr2R 14028136 113 + 21139217 GUAC---GUGUGCGGAAUUAGA----CAAGACAUUAGACUCAGAAAUCGACGGGCUUAAUGGUGAUGAACCGGGAGAGAAUGAAGGCCACCAGAAUGGUGGCGCACACGAAACAGAAGGC ...(---((((((.........----.............(((....((..(.((.(((.......))).)).)..))...)))..((((((.....)))))))))))))........... ( -33.00, z-score = -3.10, R) >droEre2.scaffold_4845 6610015 113 + 22589142 GUAC---GUGUGCGGAAUUAGA----CAAGACAUUUUGCUCAGAAAUCGACGGGCUUGAUGGUGAUGAACCGGGAGAGAAUGAAUGCCACCAGAAUGGUGGCGCACACGAAACAGAAGGC ...(---((((((.........----.....((((((.(((....(((((.....)))))(((.....)))..)))))))))...((((((.....)))))))))))))........... ( -37.70, z-score = -3.75, R) >droAna3.scaffold_13266 17153264 115 - 19884421 GUACGUGGUGUGCAGAAUUUGA----CAA-ACUUUUGGCUUAAAAGUCAACGGGCUUAAUGGUGAUAAAACGGGAGAGAAUAAAGGCCACCAGAAAGGUGGCGCACACGAAGCAAAAGGC ....((.((((((.........----...-.((.((((((....)))))).)).(((..(.((......)).)..))).......((((((.....))))))))))))...))....... ( -31.50, z-score = -2.18, R) >dp4.chr3 8156267 117 - 19779522 GUAC---GUGUGCAGAAUUUCAAUUUCGGGACUAACAGUUCAGAAGUCGACUGGCUUGAUGGUGAUAAACCGCGAGAGAAUAAAAGCCACCAGUAUGGUGGCACAGACGAAGCAAAAGGC ...(---...(((..........((((.((((.....)))).))))(((.(((.((((.((((.....)))))))).........((((((.....)))))).))).))).)))...).. ( -34.90, z-score = -2.53, R) >droPer1.super_4 2033767 117 - 7162766 GUAC---GUGUGCAGAAUUUCAAUUUCGGGACUAACAGUUCAGAAGUCGACUGGCUUGAUGGUGAUAAACCGCGAGAGAAUAAAAGCCACCAGUAUGGUGGCACAGACGAAGCAAAAGGC ...(---...(((..........((((.((((.....)))).))))(((.(((.((((.((((.....)))))))).........((((((.....)))))).))).))).)))...).. ( -34.90, z-score = -2.53, R) >droWil1.scaffold_181009 1337240 96 - 3585778 ------------------------UUUUUUUUGCUUAAUUCAAAAGUCAACCGGCUUGAUGGUGAUAAAUCGAGAGAGUAUAAAUGCCACAAAUAUUGUGGCAAUAACGAAACAAAAGGC ------------------------..(((((((....((((..(((((....)))))(((........)))....)))).....(((((((.....))))))).........))))))). ( -19.50, z-score = -1.26, R) >droMoj3.scaffold_6496 23814357 95 - 26866924 -------------------------UGUUGGCUCACAAAUCAGAAAUCAACGGGCUUGAUGGUGAUAAAGCGAGAUAAAAUGAAUGCCACCAAAAUUGUGGCACAAACGAAACAAAAGGC -------------------------((((..(((.(..((((...(((((.....)))))..))))...).)))..........((((((.......)))))).......))))...... ( -17.00, z-score = -0.06, R) >droVir3.scaffold_12875 14393447 88 + 20611582 --------------------------------UGCUGGUUCAGAAAUCAACGGGCUUGAUGGUGAUAAAGCGCGAUAGUAUAAAUGCCACCAGAACGGUGGCACAGACAAAACAGAAGGC --------------------------------(((((.............((.((((..........)))).))..........(((((((.....)))))))))).))........... ( -23.00, z-score = -1.68, R) >droGri2.scaffold_15112 4065441 100 + 5172618 ----------UGCUGGUUCA----------GUUAAUGGUUUAGAAAUCGACGGGCUUGAUGGUGAUAAAGCGCGACAGGAUAAAUGCCACCAGAACGGUGGCGCAAACGAAACAAAAGGC ----------.(((.(((..----------(((....(((((....((..((.((((..........)))).))...)).)))))((((((.....))))))...)))..)))....))) ( -24.30, z-score = -0.54, R) >consensus GUAC___GUGUGCGGAAUU__A____CAAGACUAUUGGCUCAGAAAUCGACGGGCUUGAUGGUGAUAAACCGGGAGAGAAUAAAGGCCACCAGAAUGGUGGCGCACACGAAACAAAAGGC .......................................(((...(((((.....)))))..)))....................((((((.....)))))).................. (-14.13 = -13.61 + -0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:24 2011