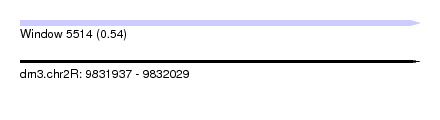
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,831,937 – 9,832,029 |
| Length | 92 |
| Max. P | 0.538132 |

| Location | 9,831,937 – 9,832,029 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.47 |
| Shannon entropy | 0.57104 |
| G+C content | 0.27555 |
| Mean single sequence MFE | -19.09 |
| Consensus MFE | -4.40 |
| Energy contribution | -4.96 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
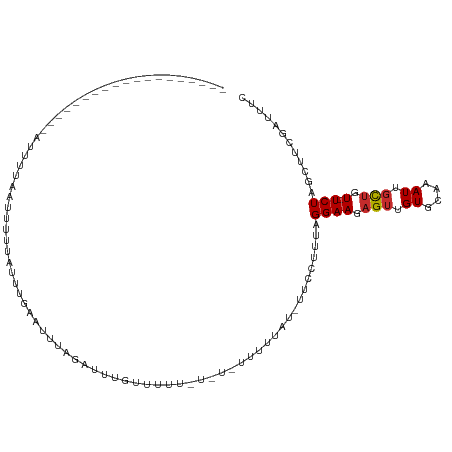
>dm3.chr2R 9831937 92 + 21146708 -------------------AAUUUAAUCGUUAGUUAGAUUUAGAUUUGUAUUUUUAUCUUUUUAU-UUCCUUUAGGAAGAGUUGUGCAAAUUGCUGU-UCUAGCUUCGAUUUC -------------------.....(((((..(((((((....(((((((((.....((((((((.-......))))))))...))))))))).....-))))))).))))).. ( -25.70, z-score = -5.22, R) >droSim1.chr2R 8265391 91 + 19596830 -------------------AAUUUAAUCGUUAGUUAGAUUUAGAUUUGUAUUUAU-UCUUUUUAU-UUCCUUUAGGAAGAGUUGUGCAAAUUGCUGU-UCUAGCUUCGAUUUC -------------------.....(((((..(((((((....(((((((((..((-((((((((.-......)))))))))).))))))))).....-))))))).))))).. ( -27.90, z-score = -6.11, R) >droSec1.super_1 7356380 91 + 14215200 ------------------AAUUUAAUCGUUAGUUAGA-UUUAGAUUUGUAUUUAU-UCUUUUUAU-UUCCUUUAGGAAGAGUUGUGCAAAUUGCUGU-UCUAGCUUCGAUUUC ------------------.....(((((..(((((((-....(((((((((..((-((((((((.-......)))))))))).))))))))).....-))))))).))))).. ( -27.90, z-score = -6.11, R) >droYak2.chr2R 14015368 105 - 21139217 AAUUAAAUCGUUACGUUAGAUUUAGAUUUAGAUUUGUAUUUGUAUUUACUUU------UUUUUAU-UUCCUUUAGGAAGAGUUGUGCAAAUUGCUGU-UCUAGCUUCGAUUUC ....((((((....((((((..(((.....(((((((((........(((((------((((...-.......))))))))).))))))))).))).-))))))..)))))). ( -21.60, z-score = -2.46, R) >droEre2.scaffold_4845 6598943 105 - 22589142 AAUUUAAUCGUUACGUUAGAUUUAGAUUUAGAUUUGUAUUUGUAUUUACUUU------UUUUUAU-UUCCUUUAGGAAGAGUUGUGCAAAUUGCUGU-UCUAGCUUCGAUUUC .....(((((....((((((..(((.....(((((((((........(((((------((((...-.......))))))))).))))))))).))).-))))))..))))).. ( -20.70, z-score = -2.27, R) >droAna3.scaffold_13266 17142572 81 + 19884421 -------------------AUUUUAAUCGUUA----GAUUUAGAUUUGUAUUUUU-------UAU-UUCCUUUAGGAAGAGUUGUGCAAAUUGCUGU-UCUAGCUUCGAUUUC -------------------.........((((----((....(((((((((..((-------(..-((((....)))))))..))))))))).....-))))))......... ( -19.10, z-score = -3.27, R) >dp4.chr3 8144584 90 + 19779522 ------------------GAUUUUGUCUUGUAUUGUAUUUUAGACUUGCUAUA---UAUUUUUAU-UUCCUUUAGGAAGAGUUGUGCAAAUUGCUGU-UCUAGCUUCGAUUUU ------------------......((((.............))))..((((..---........(-((((....)))))......((.....))...-..))))......... ( -10.22, z-score = 1.14, R) >droPer1.super_4 2023338 90 + 7162766 ------------------GAUUUUGUCUUGUAUUGUAUUUUAGACUUGCUAUA---UAUUUUUAU-UUCCUUUAGGAAGAGUUGUGCAAAUUGCUGU-UCUAGCUUCGAUUUU ------------------......((((.............))))..((((..---........(-((((....)))))......((.....))...-..))))......... ( -10.22, z-score = 1.14, R) >droWil1.scaffold_181009 1322624 91 + 3585778 -------------------GUUUUUGUUGUUACUA-AGAUUAAUUUUAUUUCCCUUUUUCAUCAUAUUCAUCUAGGAAGAGUUGUGCAAAUUUUGAUCUCUAUGUUUAGUA-- -------------------...........(((((-(((((((..((......(((((((..............))))))).......))..))))))).......)))))-- ( -8.47, z-score = 1.00, R) >consensus ___________________AUUUUAAUUUUUAUUUGAAUUUAGAUUUGUUUUU_U_U_UUUUUAU_UUCCUUUAGGAAGAGUUGUGCAAAUUGCUGU_UCUAGCUUCGAUUUC ..................................................................((((....))))((((((.(((......)))...))))))....... ( -4.40 = -4.96 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:19 2011