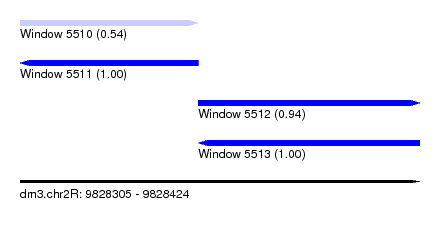
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,828,305 – 9,828,424 |
| Length | 119 |
| Max. P | 0.998718 |

| Location | 9,828,305 – 9,828,358 |
|---|---|
| Length | 53 |
| Sequences | 5 |
| Columns | 55 |
| Reading direction | forward |
| Mean pairwise identity | 94.24 |
| Shannon entropy | 0.10368 |
| G+C content | 0.56403 |
| Mean single sequence MFE | -18.92 |
| Consensus MFE | -16.54 |
| Energy contribution | -16.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
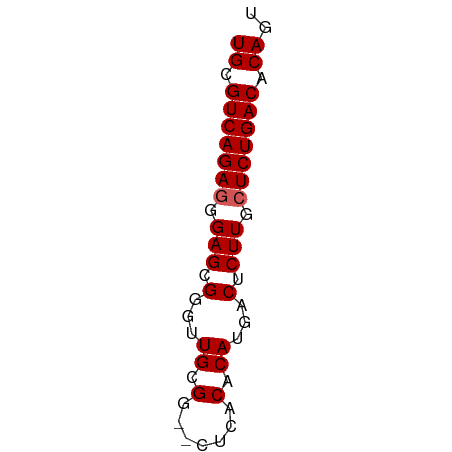
>dm3.chr2R 9828305 53 + 21146708 UGCGUCAGAGGGAGCGGGUUGCGG--CUCACACAUGACUCUUGCUCUGACACAGU ((.(((((((.(((.(...((.(.--....).))...).))).))))))).)).. ( -18.90, z-score = -1.15, R) >droSim1.chr2R 8261689 53 + 19596830 UGCGUCAGAGGGAGCGGGUUGCGG--CUCACACAUGACUCUUGCUCUGACACAGU ((.(((((((.(((.(...((.(.--....).))...).))).))))))).)).. ( -18.90, z-score = -1.15, R) >droSec1.super_1 7352809 53 + 14215200 UGCGUCAGAGGGAGCGGAUUGCGA--CUCACACAUGACUCUUGCUCUGACACAGU ((.(((((((.(((.(...((.(.--....).))...).))).))))))).)).. ( -17.60, z-score = -1.60, R) >droYak2.chr2R 14011654 55 - 21139217 UGCGUCAGAGGGAGCGGGUUGUGGAUCACACACAUGACUCUUGCUCUGACACAGU ((.(((((((.(((.(...((((.......))))...).))).))))))).)).. ( -22.50, z-score = -2.69, R) >droEre2.scaffold_4845 6595448 52 - 22589142 UGCGUCAGAGGGAGCGGGUUGCG---CUCACACAUGACUCUUGAUCUGACACAGU ((.((((((..(((((.....))---)))...((.......)).)))))).)).. ( -16.70, z-score = -0.99, R) >consensus UGCGUCAGAGGGAGCGGGUUGCGG__CUCACACAUGACUCUUGCUCUGACACAGU ((.(((((((.(((.(...((.(.......).))...).))).))))))).)).. (-16.54 = -16.74 + 0.20)

| Location | 9,828,305 – 9,828,358 |
|---|---|
| Length | 53 |
| Sequences | 5 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 94.24 |
| Shannon entropy | 0.10368 |
| G+C content | 0.56403 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -21.80 |
| Energy contribution | -21.84 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.16 |
| Structure conservation index | 1.05 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.998718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
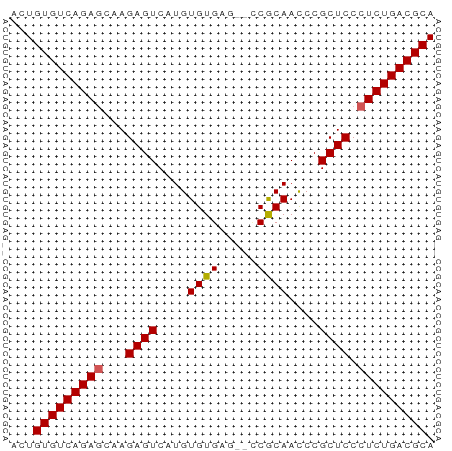
>dm3.chr2R 9828305 53 - 21146708 ACUGUGUCAGAGCAAGAGUCAUGUGUGAG--CCGCAACCCGCUCCCUCUGACGCA ...(((((((((...((((....((((..--.))))....)))).))))))))). ( -20.90, z-score = -3.09, R) >droSim1.chr2R 8261689 53 - 19596830 ACUGUGUCAGAGCAAGAGUCAUGUGUGAG--CCGCAACCCGCUCCCUCUGACGCA ...(((((((((...((((....((((..--.))))....)))).))))))))). ( -20.90, z-score = -3.09, R) >droSec1.super_1 7352809 53 - 14215200 ACUGUGUCAGAGCAAGAGUCAUGUGUGAG--UCGCAAUCCGCUCCCUCUGACGCA ...(((((((((...((((....((((..--.))))....)))).))))))))). ( -20.90, z-score = -3.13, R) >droYak2.chr2R 14011654 55 + 21139217 ACUGUGUCAGAGCAAGAGUCAUGUGUGUGAUCCACAACCCGCUCCCUCUGACGCA ...(((((((((...((((....((((.....))))....)))).))))))))). ( -22.40, z-score = -3.96, R) >droEre2.scaffold_4845 6595448 52 + 22589142 ACUGUGUCAGAUCAAGAGUCAUGUGUGAG---CGCAACCCGCUCCCUCUGACGCA ...((((((((....((((..((((....---))))....))))..)))))))). ( -18.30, z-score = -2.51, R) >consensus ACUGUGUCAGAGCAAGAGUCAUGUGUGAG__CCGCAACCCGCUCCCUCUGACGCA ...(((((((((...((((....((((.....))))....)))).))))))))). (-21.80 = -21.84 + 0.04)

| Location | 9,828,358 – 9,828,424 |
|---|---|
| Length | 66 |
| Sequences | 8 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 81.49 |
| Shannon entropy | 0.33653 |
| G+C content | 0.39978 |
| Mean single sequence MFE | -15.60 |
| Consensus MFE | -9.62 |
| Energy contribution | -9.65 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
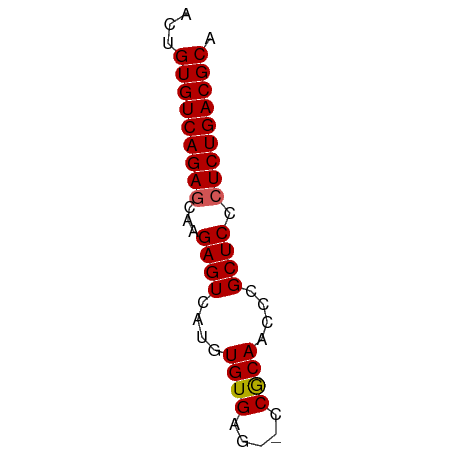
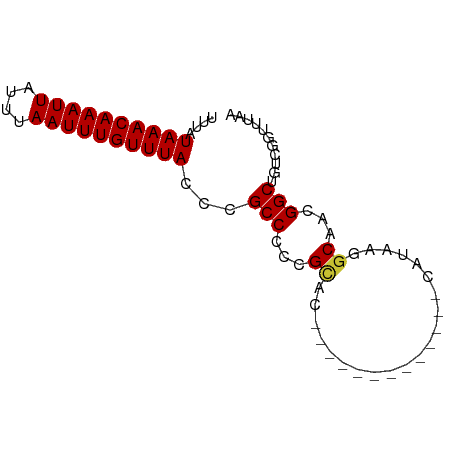
>dm3.chr2R 9828358 66 + 21146708 UUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGUAC-------------AAUAAAGCAACGGCUGUCGGUUUAA ....((((((((((....)))))))))).((((..((((..-------------.........))))..).)))..... ( -13.20, z-score = -1.83, R) >droPer1.super_4 2014669 70 + 7162766 UUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCUCGCCUCCA---------ACAGAAGGCAGCGGCUGUUCCACACA ....((((((((((....))))))))))...(((((.((((.(.---------...).)))))).)))........... ( -18.10, z-score = -4.08, R) >dp4.chr3 8100347 70 + 19779522 UUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCUCGACUCAA---------ACAGAAGGCAGCGGCUGUUCCACACA ....((((((((((....)))))))))).((((((((.......---------...).)))..))))............ ( -13.10, z-score = -2.05, R) >droAna3.scaffold_13266 17130065 75 + 19884421 UUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCCUCCACACUUCACAACAUAAGGCCACGGCUGUUUAA---- ....((((((((((....))))))))))...(((...((((.................))))...))).......---- ( -14.53, z-score = -3.44, R) >droEre2.scaffold_4845 6595500 66 - 22589142 UUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCAG-------------CAAAAGGCCGCGGCUGUCGGUUUAA ....((((((((((....)))))))))).((((..((((.(-------------(.....)).))))..).)))..... ( -19.30, z-score = -2.47, R) >droYak2.chr2R 14011709 66 - 21139217 UUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCAG-------------CAUAAGGCAGCGGCUGUCGGUUUAA ....((((((((((....)))))))))).((((..((((.(-------------(.....)).))))..).)))..... ( -18.60, z-score = -2.47, R) >droSec1.super_1 7352862 66 + 14215200 UUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCAC-------------CAUAAGGCAACGGCUGUCGGUUUAA ....((((((((((....)))))))))).((((..(((..(-------------(....))...)))..).)))..... ( -14.00, z-score = -1.41, R) >droSim1.chr2R 8261742 66 + 19596830 UUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCAC-------------CAUAAGGCAACGGCUGUCGGUUUAA ....((((((((((....)))))))))).((((..(((..(-------------(....))...)))..).)))..... ( -14.00, z-score = -1.41, R) >consensus UUUAUAAACAAAUUAUUUAAUUUGUUUACCCGCCCCCGCAC_____________CAUAAGGCAACGGCUGUCGGUUUAA ....((((((((((....))))))))))...(((...((.....................))...)))........... ( -9.62 = -9.65 + 0.03)

| Location | 9,828,358 – 9,828,424 |
|---|---|
| Length | 66 |
| Sequences | 8 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 81.49 |
| Shannon entropy | 0.33653 |
| G+C content | 0.39978 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.24 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
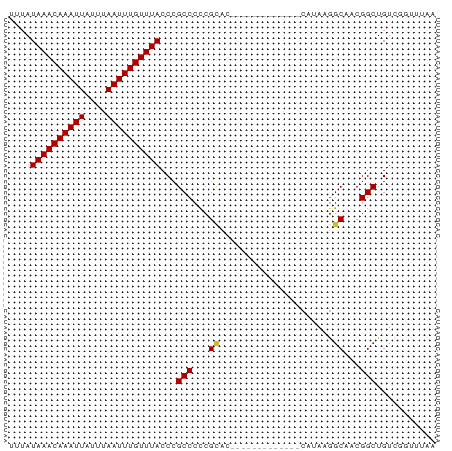
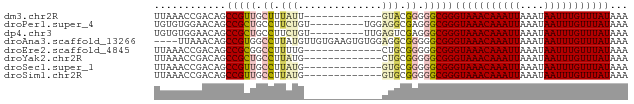
>dm3.chr2R 9828358 66 - 21146708 UUAAACCGACAGCCGUUGCUUUAUU-------------GUACGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAA .....(((.(..((((.((......-------------))))))..))))(((((((((((....)))))))))))... ( -19.40, z-score = -3.06, R) >droPer1.super_4 2014669 70 - 7162766 UGUGUGGAACAGCCGCUGCCUUCUGU---------UGGAGGCGAGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAA (((.....))).(((((((((((...---------.))))))...)))))(((((((((((....)))))))))))... ( -25.50, z-score = -3.95, R) >dp4.chr3 8100347 70 - 19779522 UGUGUGGAACAGCCGCUGCCUUCUGU---------UUGAGUCGAGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAA ...((((.....))))(((((((...---------.......))))))).(((((((((((....)))))))))))... ( -20.70, z-score = -2.64, R) >droAna3.scaffold_13266 17130065 75 - 19884421 ----UUAAACAGCCGUGGCCUUAUGUUGUGAAGUGUGGAGGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAA ----.......(((.(.((((((..((....))..).))))).).)))..(((((((((((....)))))))))))... ( -24.50, z-score = -3.90, R) >droEre2.scaffold_4845 6595500 66 + 22589142 UUAAACCGACAGCCGCGGCCUUUUG-------------CUGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAA .....(((.(..(((((((.....)-------------))))))..))))(((((((((((....)))))))))))... ( -26.40, z-score = -4.03, R) >droYak2.chr2R 14011709 66 + 21139217 UUAAACCGACAGCCGCUGCCUUAUG-------------CUGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAA .....(((.(..((((.((.....)-------------).))))..))))(((((((((((....)))))))))))... ( -22.50, z-score = -3.02, R) >droSec1.super_1 7352862 66 - 14215200 UUAAACCGACAGCCGUUGCCUUAUG-------------GUGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAA .....(((.(..((((.(((....)-------------))))))..))))(((((((((((....)))))))))))... ( -21.50, z-score = -2.77, R) >droSim1.chr2R 8261742 66 - 19596830 UUAAACCGACAGCCGUUGCCUUAUG-------------GUGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAA .....(((.(..((((.(((....)-------------))))))..))))(((((((((((....)))))))))))... ( -21.50, z-score = -2.77, R) >consensus UUAAACCGACAGCCGCUGCCUUAUG_____________AUGCGGGGGCGGGUAAACAAAUUAAAUAAUUUGUUUAUAAA ............(((((.((......................)).)))))(((((((((((....)))))))))))... (-15.10 = -15.24 + 0.14)
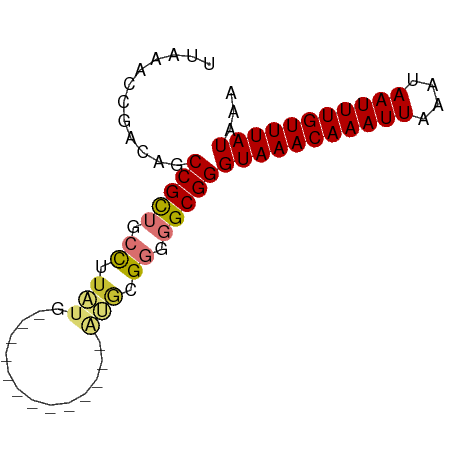
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:18 2011