
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,798,672 – 2,798,764 |
| Length | 92 |
| Max. P | 0.755823 |

| Location | 2,798,672 – 2,798,764 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Shannon entropy | 0.34661 |
| G+C content | 0.57274 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.755823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2798672 92 - 23011544 UAAAUCAG---GCAGCAGCAGCAGC----ACACAUACAUCGAC-AAGCGCCAGGACCCGCAUCACCAU-AUCCUUCCUGCCAAAUAGUCGCCACUGCUGCC .......(---(((((((..((.((----..............-..))(.(((((.............-.....))))).)........))..)))))))) ( -22.86, z-score = -1.97, R) >droSim1.chr2L 2756359 94 - 22036055 UAAAUCAG---GCAGCAGCAGCAGCU--CACACAUACAUCGAC-AAGCGCCAGGAUCCGCAUCACCAU-AUCCUUCCUGCCAAAUAGUCGCCACUGCUGCC .......(---(((((((..((.(((--...............-.)))(.(((((.............-.....))))).)........))..)))))))) ( -24.56, z-score = -2.42, R) >droSec1.super_5 957971 97 - 5866729 UAAAUCAGGCAGCAGCAGCAGCAGCU--CACACAUACAUCGAC-AAGCGCCAGGAUCCGCAUCACCAU-AUCCUUCCUGCCAAAUAGUCGCCACUGCUGCC .......((((((((..((.((.(((--...............-.)))(.(((((.............-.....))))).).....)).))..)))))))) ( -25.86, z-score = -2.39, R) >droYak2.chr2L 2790754 100 - 22324452 AAUAACAGGCAGCAGCAGGAGGAGCUCACACACACACAUCGACGAAGCGCCAGGACCCGCAUCACCAU-AUCCUUCCUGCCAAACAGUCGCCACUGCUGCC .......((((((.((((((((((......)............((.(((........))).)).....-.))).)))))).....(((....))))))))) ( -27.00, z-score = -1.79, R) >droEre2.scaffold_4929 2845102 99 - 26641161 AAUAGCAGGCAGCAGCAGGAGCAGCUCACACACACACAUCGAC-AAGCGCCAGGACCCGCAUCACCAU-AUCCUUCCUGCCAAACAGUCGCCACUGCUGCC .......((((((.((((((((.(((.................-.))))).((((.............-.)))))))))).....(((....))))))))) ( -23.61, z-score = -0.61, R) >dp4.chr4_group4 2138235 80 - 6586962 -----------GCAGCAGCAGGAGCA-ACUCACACACACCGA---------AGGACCCGCAUCAUCAUCAUCUUUCCUGCCCAGUCGUCGCCACUGCUGCC -----------...(((((((..((.-((..((.......((---------((((...............)))))).......)).)).))..))))))). ( -19.60, z-score = -1.19, R) >droPer1.super_10 1147747 80 - 3432795 -----------GCAGCAGCAGGAGCA-ACUCACACACACCGA---------AGGACCCGCAUCAUCAUCAUCUUUCCUGCCCAGUCGUCGCCACUGCUGCC -----------...(((((((..((.-((..((.......((---------((((...............)))))).......)).)).))..))))))). ( -19.60, z-score = -1.19, R) >consensus UAAAUCAG___GCAGCAGCAGCAGCU_ACACACACACAUCGAC_AAGCGCCAGGACCCGCAUCACCAU_AUCCUUCCUGCCAAAUAGUCGCCACUGCUGCC ...........(((((((.....(......)........((((.......(((((...................))))).......))))...))))))). (-15.78 = -16.35 + 0.57)
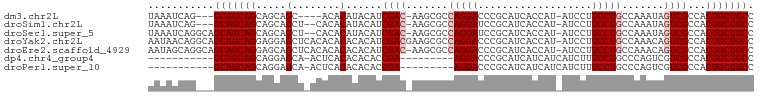
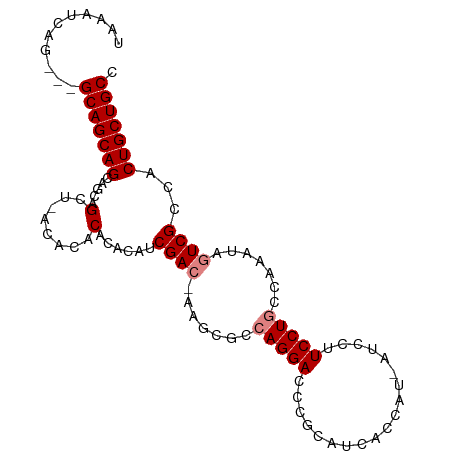

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:17 2011