
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,795,300 – 9,795,393 |
| Length | 93 |
| Max. P | 0.592460 |

| Location | 9,795,300 – 9,795,393 |
|---|---|
| Length | 93 |
| Sequences | 14 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 62.80 |
| Shannon entropy | 0.81054 |
| G+C content | 0.58639 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -9.66 |
| Energy contribution | -9.92 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.592460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
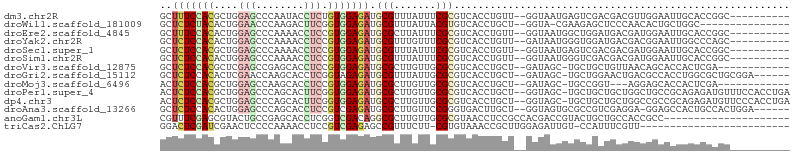
>dm3.chr2R 9795300 93 + 21146708 GCUUUCCACGCUGGAGCCCAAUACCUCUGUGGAGAUGCGUUUAUUUCGCGUCACCUGUU--GGUAAUGAGUCGACGACGUUGGAAUUGCACCGGC---------- ((.(((((((.((..((.((.((((.....((.((((((.......)))))).))....--)))).)).).)..)).)).)))))..))......---------- ( -29.10, z-score = -0.98, R) >droWil1.scaffold_181009 2169663 87 - 3585778 GCUCUCUACACUGGAACCCAAGACUUCGGUGGAGAUGCGUUUAUUACGUGUCACCUGCU--GGUA-CGAAGAGCUCCCAACACUGCUGGC--------------- ((((((..(((((((.........)))))))..(((((((.....)))))))(((....--))).-.).)))))..(((.......))).--------------- ( -23.60, z-score = -0.32, R) >droEre2.scaffold_4845 6562714 93 - 22589142 GCUUUCCACACUGGAGCCCAAAACCUCCGUGGAGAUGCGCUUAUUUCGCGUCACCUGUU--GGUAAUGGCUGGAUGACGAUGGAAUUGCACCGGC---------- (((((((((...((((........))))))))))).))((..(((((((((((((.(((--......))).)).))))).)))))).))......---------- ( -32.00, z-score = -1.83, R) >droYak2.chr2R 13978806 93 - 21139217 GCUCUCCACACUGGAGCCCAAAACCUCCGUGGAGAUGCGUUUGUUUCGCGUCACCUGUU--GAUAAUGGGUGGAUGACGACGGAAUUGGCCCAGC---------- (((((((((...((((........))))))))))).))((..((((((((((((((((.--....))))))....)))).))))))..)).....---------- ( -33.70, z-score = -1.90, R) >droSec1.super_1 7320271 93 + 14215200 GCUCUCCACGCUGGAGCCCAAAACCUCCGUGGAGAUGCGUUUAUUUCGCGUCACCUGUU--GGUAAUGAGUCGACGACGAUGGAAUUGCACCGGC---------- ((((((((((..((.........))..)))))))).))((..(((((((((((((....--))).....(....))))).)))))).))......---------- ( -29.70, z-score = -0.96, R) >droSim1.chr2R 8227293 93 + 19596830 GCUCUCCACACUGGAGCCCAAAACCUCCGUGGAGAUGCGUUUAUUUCGCGUCACCUGUU--GGUAAUGGGUCGACGACGAUGGAAUUGCACCGGC---------- ..(((((((...((((........)))))))))))(((((((...(((((((((((((.--....)))))).)))).)))..))).)))).....---------- ( -32.60, z-score = -1.61, R) >droVir3.scaffold_12875 14333582 90 - 20611582 GCUCUCCACGCUCGAGCCGAGCACCUCCGUGGAGAUGCGCUUGUUGCGCGUCACCUGCU--GAUAGC-UGCUGCUGUUAACAGCACCACUCGA------------ .........(((((...)))))......((((.(((((((.....)))))))..(((.(--((((((-....))))))).)))..))))....------------ ( -35.80, z-score = -2.29, R) >droGri2.scaffold_15112 4006493 96 - 5172618 GCUCUCCACACUCGAACCAAGCACCUCGGUAGAGAUGCGUUUAUUGCGCGUCACCUGCU--GAUAGC-UGCUGGAACUGACGCCACCUGGCGCUGCGGA------ ....(((.((......(((.(((.((((((((.(((((((.....)))))))..)))))--))..).-))))))......((((....)))).)).)))------ ( -33.90, z-score = -1.63, R) >droMoj3.scaffold_6496 23752081 87 + 26866924 ACUCUCCACGCUGGAGCCAAGCACCUCCGUGGAGAUGCGCUUGUUGCGCGUCACCUGCU--GAUAGC-UGCCGGU---AGGAGCACCACUCGA------------ ....((((((..((.((...)).))..))))))(((((((.....))))))).((((((--(.....-...))))---)))............------------ ( -35.70, z-score = -1.73, R) >droPer1.super_4 1983242 102 + 7162766 ACUCUCCACGCUGGAGCCCAGCACUUCGGUGGAGAUGCGCUUGUUGCGCGUCACCUGCU--GGUAGC-UGCUGCUGCUGGCUGCCGCAGAGAUGUUUCCACCUGA .........(((((...))))).....(((((((((((((.....)))))))..((((.--((((((-(((....)).))))))))))).......))))))... ( -50.00, z-score = -3.37, R) >dp4.chr3 8069697 102 + 19779522 ACUCUCCACGCUGGAGCCCAGCACUUCGGUGGAGAUGCGCUUGUUGCGCGUCACCUGCU--GGUAGC-UGCUGCUGCUGGCCGCCGCAGAGAUGUUCCCACCUGA .........(((((...))))).....(((((.(((((((.....)))))))..((((.--((..((-(((....)).)))..))))))........)))))... ( -44.80, z-score = -1.73, R) >droAna3.scaffold_13266 17096147 96 + 19884421 GCUCUCCACACUGGAGCCCAGCACCUCCGUCGAGAUGCGCUUGUUCCGGGUGACUUGCU--GGUAGUGCGCCGUCGAGGA-GGAGCCACUGCCACUGGA------ ((....(((.(((((((...(((.(((....))).)))....))))))))))....))(--(((((((.(((.((...))-.).)))))))))).....------ ( -37.20, z-score = -0.01, R) >anoGam1.chr3L 22806601 84 + 41284009 CGUUUCGAGCGUACUGCCGAGCACCUCGGUCGACAGGCGCUUGUUGCGCGUAACCUCCGCCACGACCGUACUGCUGCCACCGCC--------------------- ........(((....((..((((...((((((...((((...((((....))))...)))).))))))...))))))...))).--------------------- ( -26.90, z-score = 0.16, R) >triCas2.ChLG7 7163089 78 + 17478683 GGACUCGAUCGAACUCCCCAAAACCUCCGUCGAGAGCCGUUUCUU-CGUGUAAACCGCUUGGAGAUUGU-CCAUUUCGUU------------------------- ((((((....)).((((.......(((....)))(((.(((((..-...).)))).))).))))...))-))........------------------------- ( -16.30, z-score = 0.22, R) >consensus GCUCUCCACACUGGAGCCCAACACCUCCGUGGAGAUGCGCUUGUUGCGCGUCACCUGCU__GGUAGCGUGCCGAUGACGACGGAACCGCACCGGC__________ ..(((((((....(((........))).))))))).(((.......)))........................................................ ( -9.66 = -9.92 + 0.26)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:10 2011