
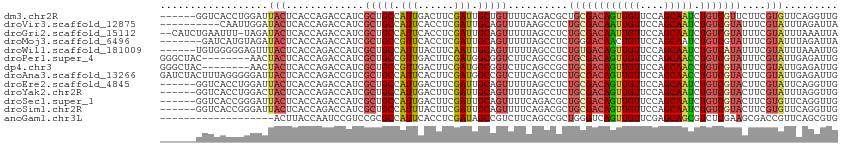
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,786,935 – 9,787,042 |
| Length | 107 |
| Max. P | 0.871143 |

| Location | 9,786,935 – 9,787,042 |
|---|---|
| Length | 107 |
| Sequences | 13 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.07 |
| Shannon entropy | 0.49140 |
| G+C content | 0.48868 |
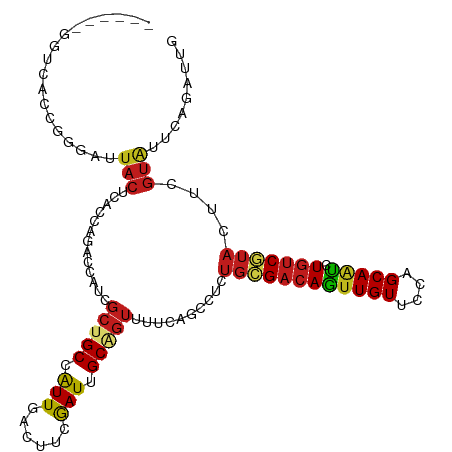
| Mean single sequence MFE | -28.59 |
| Consensus MFE | -15.76 |
| Energy contribution | -15.84 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9786935 107 + 21146708 ------GGUCACCUGGAUUACUCACCAGACCAUCGCUGCCAUUGACUUCGAUUGCUGUUUUCAGACGCUGCGACAGUUGUUCCAGCAAUCUGUCGUUCUUCGUGUUCAGGUUG ------....((((((((.((.............((.((.((((....)))).)).)).....((.(..(((((((((((....)))).))))))).).)))))))))))).. ( -28.40, z-score = -0.49, R) >droVir3.scaffold_12875 14326335 103 - 20611582 ----------CAAUUGGAUACUCACCAGACCAUCGCUGCCAUUCACCUCGAUUGCAGUUUUAAGCCUCUGCGACAAUUGUUCCAGCAAUCUGUCGUAUUUCGUAUUUAGAUUA ----------...((((((((.............(((((.(((......))).)))))..........((((((((((((....))))).)))))))....)))))))).... ( -23.00, z-score = -2.10, R) >droGri2.scaffold_15112 3998702 110 - 5172618 --CAUCUGAAUUU-UAGAUACUCACCAGACCAUCGCUGCCAUUCACCUCGAUUGCAGUUUUUAGCCUCUGCGACAAUUGUUCCAGCAAUCUGUCGUAUUUCGUAUUUAAAUUA --.........((-(((((((.............(((((.(((......))).)))))..........((((((((((((....))))).)))))))....)))))))))... ( -24.30, z-score = -3.16, R) >droMoj3.scaffold_6496 23744544 106 + 26866924 -------GAUCAUGUAGAUACUCACCAGACCAUCGCUGCCGUUCACCUCGAUUGCAGUUUUUAGCCUCUGGGACAACUGUUCCAGCAAUCUGUCGUAUUUCGUAUUUAGAUUA -------....((.(((((((......(((....(((((.(((......))).))))).........(((((((....)))))))......))).......))))))).)).. ( -23.32, z-score = -1.03, R) >droWil1.scaffold_181009 2163383 107 - 3585778 ------UGUGGGGGAGUUUACUCACCAGACCAUCGCUGCCAUUUACUUCAAUUGCAGUUUUUAGCCUCUGUGACAGUUGUUCCAGCAAUCUGUCAUAUUUCGUAUUUAAAUUG ------...(((((((....)))...........(((((.(((......))).)))))......))))((((((((((((....)))).))))))))................ ( -25.50, z-score = -1.50, R) >droPer1.super_4 1976636 105 + 7162766 GGGCUAC--------AACUACUCACCAGACCAUCGCUGCCGUUGACUUCGAUGGCGGUCUUCAGCCGCUGCGACAGUUGUUCCAGCAACCUGUCGUAUUUCGUAUUGAGAUUG .((((..--------......((....)).....((((((((((....))))))))))....))))..((((((((((((....)))).))))))))((((.....))))... ( -33.40, z-score = -1.79, R) >dp4.chr3 8063077 105 + 19779522 GGGCUAC--------AACUACUCACCAGACCAUCGCUGCCGUUGACUUCGAUGGCGGUCUUCAGCCGCUGCGACAGUUGUUCCAGCAACCUGUCGUAUUUCGUAUUGAGAUUG .((((..--------......((....)).....((((((((((....))))))))))....))))..((((((((((((....)))).))))))))((((.....))))... ( -33.40, z-score = -1.79, R) >droAna3.scaffold_13266 17089685 113 + 19884421 GAUCUACUUUAGGGGGAUUACUCACCAGACCGUCGCUGCCAUUCACUUCGAUGGCCGUCUUCAGCCUCUGCGACAGUUGUUCCAGCAACCUGUCGUACUUCGUAUUGAGAUUG (((((.(..((.((((...............(..(((((((((......)))))).......)))..)((((((((((((....)))).)))))))))))).))..)))))). ( -30.61, z-score = -0.82, R) >droEre2.scaffold_4845 6556315 107 - 22589142 ------GGUCACCUGGAUUACUCACCAGACCAUCGCUGCCAUUGACUUCGAUUGCAGUUUUUAGCCUCUGCGACAGUUGUUCCAGCAAUCUGUCGUACUUCGUAUUCAGGUUG ------....((((((((.((.............(((((.((((....)))).)))))..........((((((((((((....)))).))))))))....)))))))))).. ( -33.00, z-score = -3.17, R) >droYak2.chr2R 13972403 107 - 21139217 ------GGUCACCUGGACUACUCACCAGACCAUCGCUGCCAUUGACUUCGAUUGCAGUUUUUAGCCUCUGCGACAGUUGUUCCAGCAAUCUGUCGUACUUCGUAUUUAGGUUG ------((((...(((........)))))))...(((((.((((....)))).)))))...((((((.((((((((((((....)))).))))))))..........)))))) ( -31.80, z-score = -2.74, R) >droSec1.super_1 7313842 107 + 14215200 ------GGUCACCGGGAUUACUCACCAGACCAUCGCUGCCAUUGACUUCGAUUGCAGUUUUCAGACGCUGCGACAGUUGUUCCAGCAAUCUGUCGUACUUCGUGUUCAGGUUG ------((((...((((....)).)).))))((((((((.((((....)))).))))).....(((((((((((((((((....)))).))))))))....)))))..))).. ( -32.90, z-score = -1.60, R) >droSim1.chr2R 8220861 107 + 19596830 ------GGUCACCGGGAUUACUCACCAGACCAUCGCUGCCAUUUACUUCGAUUGCAGUUUUCAGACGCUGCGACAGUUGUUCCAGCAAUCUGUCGUACUUCGUGUUCAGGUUG ------((((...((((....)).)).))))((((((((.(((......))).))))).....(((((((((((((((((....)))).))))))))....)))))..))).. ( -29.70, z-score = -1.28, R) >anoGam1.chr3L 22792453 94 + 41284009 -------------------ACUUACCAAUCCGUCCGCGCCAUUCACCUCGAUAGCCGUCUUCAGCCGCUGGGUCAGUUGUUCGAGCAGCGUCUCGAAGCGACCGUUCAGCGUG -------------------...............(((((............((((.((.....)).))))((((.((..((((((......)))))))))))).....))))) ( -22.40, z-score = 0.75, R) >consensus ______GGUCACCGGGAUUACUCACCAGACCAUCGCUGCCAUUGACUUCGAUUGCAGUUUUCAGCCUCUGCGACAGUUGUUCCAGCAAUCUGUCGUACUUCGUAUUCAGAUUG ..................(((.............(((((.(((......))).)))))..........((((((((((((....))))).)))))))....)))......... (-15.76 = -15.84 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:10 2011