
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,761,712 – 9,761,823 |
| Length | 111 |
| Max. P | 0.708052 |

| Location | 9,761,712 – 9,761,823 |
|---|---|
| Length | 111 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.57 |
| Shannon entropy | 0.74750 |
| G+C content | 0.49504 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -11.51 |
| Energy contribution | -11.81 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.708052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9761712 111 + 21146708 ---------CAAGCAUGAAAUAUAUAUCUACGUACCGGCUUCAGUUGGUCCAGUGUGCUGUCGGUCUUGUUGAUCCAUACCAGCAGCUCGUUGAGAGCAUGCUGGAAUUGACCCAAGUGC ---------...((((..................(((((....)))))(((((((((((.((((..((((((........))))))....)))).)))))))))))..........)))) ( -33.60, z-score = -1.21, R) >droSim1.chr2R_random 2311633 101 + 2996586 ---------ACAGAAUUAA-UAUAUAUCUACGUACCG--CUGAGUUG-UCCA-UGUGCUGUCGGCCU-GUUGAAC-AGACCAGUAGCUUCUCAGGG--AUGCUGGAAUUAACC-AAAUGC ---------..........-......((((.(((((.--(((((...-....-...((((..((.((-(.....)-)).))..))))..)))))))--.))))))).......-...... ( -23.40, z-score = -0.04, R) >droSec1.super_1 7288604 111 + 14215200 ---------CCAGAAUUAAAUAUAUAUCUACGUACCGGCUUAAGUUGGUCCAGCGUGCUGUCGGUCUUGUUGAUCCAGACCAGUAGCUCAUUGAGGGCAUGCUGGAAUUGACCCAAGUGC ---------............................((((..((..((((((((((((.(((((((.(.....).))))).(.....)...)).)))))))))).))..))..)))).. ( -35.10, z-score = -1.92, R) >droYak2.chr2R 13947213 111 - 21139217 ---------CAAGAAUUAUAUAUAUAGUUACGUACCGGCUUCAGUUGGUCCAGAGUGCUGUCGGUCUUGUUGAUCCAGACCAGCAGCUCGUUGAGGGCAUGCUGGAAUUGACCCAAAUGC ---------.................((((....(((((....)))))(((((.(((((.(((((((.(.....).)))))(((.....))))).))))).)))))..))))........ ( -30.50, z-score = -0.42, R) >droEre2.scaffold_4845 6531131 108 - 22589142 ---------UAAGAAUUAU---UAUAGUUACGUACCGGCUUCAGUUGGUCCAGUGUGCUGUCGGUCUUGUUGAUCCAGACCAGCAGCUCGUUGAGGGCAUGCUGGAAUUGACCCAAGUGC ---------..........---...............((((..((..((((((((((((.(((((((.(.....).)))))(((.....))))).)))))))))).))..))..)))).. ( -34.20, z-score = -1.42, R) >droAna3.scaffold_13266 17064549 96 + 19884421 ------------------------UAAAAACUUACUGGUUUCAACUGAUCCAGAGUUCCAUCGGUCUUGUUGAUCCAGACCAGCAGUUCGUUGAGGGCGUGCUGGAAUUGACCCAAGUGG ------------------------.....((((.((((..(((((.((.((.((......))))))..))))).)))).((((((((((.....)))).)))))).........)))).. ( -29.90, z-score = -1.59, R) >dp4.chr3 8036841 115 + 19779522 -----CUGGGAUUCGACCCGAGCUUUAGCGCUUACCGGUUUCAACUGAUCCAAAGUGCCGUCCGUCCUGUCGAUCCAGACCAGCAGCUCGUUGAGAGCGUGCUGGAAUUGACCCAAGUGG -----.((((..(((((.((..(....((((((...((((......))))..)))))).)..))....)))))......((((((((((.....)))).))))))......))))..... ( -37.90, z-score = -1.27, R) >droPer1.super_4 1950344 115 + 7162766 -----CUGGGAUUCGACCCGAGCUUUAGCGCUUACCGGCUUCAACUGAUCCAAAGUGCCGUCCGUCCUGUCGAUCCAGACCAGCAGCUCGUUGAGAGCGUGCUGGAAUUGACCCAAGUGG -----.((((..(((((..((((......))))..((((....(((.......)))))))........)))))......((((((((((.....)))).))))))......))))..... ( -38.40, z-score = -1.19, R) >droWil1.scaffold_181009 2137541 84 - 3585778 ------------------------------------GGUUUCAGUUGAUCUAAGGUGCCAUCUGUCUUGUUGAUCCAAACGAGCAACUCGUUCAAAGCAUGUUGGAACUGUCCCAAAUGA ------------------------------------((...((((...(((((.((((.....(((.....)))...((((((...))))))....)))).)))))))))..))...... ( -19.90, z-score = -0.65, R) >droVir3.scaffold_12875 14298177 112 - 20611582 --------UUGGAAGCUACUCCUUCGCUAUCUUACCGGUUUCAACUGAUCCAAUGUGCUGUCUGUUUUGUUAAUCCAGACCAACAGCUCGUUGAGUGCAUGCUGGAAUUGUCCCAAAUGG --------((((.(((.........)))......(((((....(((....(((((.(((((.((.((((......)))).))))))).))))))))....))))).......)))).... ( -27.20, z-score = -0.97, R) >droMoj3.scaffold_6496 23716258 112 + 26866924 --------CUAGAAGCUAAAACGUGGCUCUCUUACCGGUUUCAACUGAUCCAAUGUGUUGUCUGUCUUGUUAAUCCAAACCAACAGCUCAUUGAGAGCAUGUUGGAAUUGACCCAAGUGG --------..((((((((.....))))).)))...((((((((((((.(((((((.(((((.((..(((......)))..))))))).))))).)).)).)))))))))).......... ( -30.50, z-score = -1.90, R) >droGri2.scaffold_15112 3971589 120 - 5172618 UUCAAAGAAAUAACUCUUCUUUGCUUUCUUCUUACCGGCUUCAGCUGAUCCAGAGUGCUGUCUGUCUUGUUUAUCCAGACCAACAGCUCGUUGAGCGCAUGCUGGAAUUGACCCAAGUGG ..(((((((.......))))))).........(((((((....)))).(((((.(((((((..((((.(.....).))))..)))((((...)))))))).)))))..........))). ( -30.80, z-score = -0.82, R) >anoGam1.chr3L 22757218 84 + 41284009 ------------------------------------GGUUUCAGCUCGUCCAGGUUGGCGUCGGUCUUGUUGAUCCACACGAGCAGCUCGUUCAGCGCGUGCUGGAACUGCCCGAGAUGC ------------------------------------.(((((.((((((...(((..(((.......)))..)))...)))))).((...((((((....))))))...))..))))).. ( -29.70, z-score = -0.13, R) >triCas2.ChLG7 7134025 84 + 17478683 ------------------------------------GGUUUCAGCUCAUCCAAAGUCUUAUCAGUCUUGCUGAUCCAGACCAGGAGCUCGUUGAGGGCAUGUUGGAACUGGCCGAGACGC ------------------------------------.(((((.(((..(((((.((((.(((((.....)))))..)))).....((((.....))))...)))))...))).))))).. ( -28.00, z-score = -0.96, R) >consensus __________AAGAAUUA____UAUAUCUACUUACCGGUUUCAGCUGAUCCAGAGUGCUGUCGGUCUUGUUGAUCCAGACCAGCAGCUCGUUGAGGGCAUGCUGGAAUUGACCCAAGUGC ....................................((((......)))).............................((((((((((.....)))).))))))............... (-11.51 = -11.81 + 0.30)

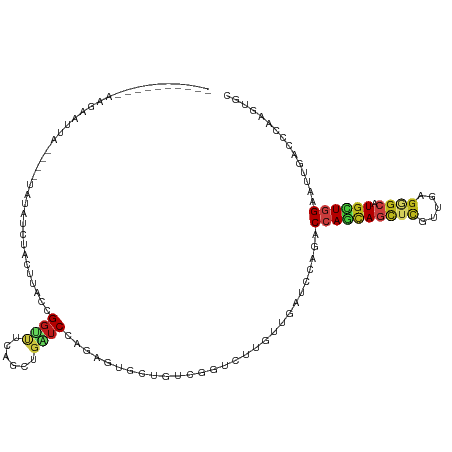
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:05 2011