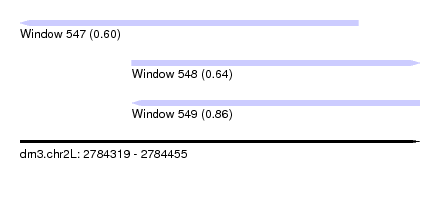
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,784,319 – 2,784,455 |
| Length | 136 |
| Max. P | 0.859345 |

| Location | 2,784,319 – 2,784,434 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.59 |
| Shannon entropy | 0.38594 |
| G+C content | 0.42947 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -15.47 |
| Energy contribution | -15.91 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
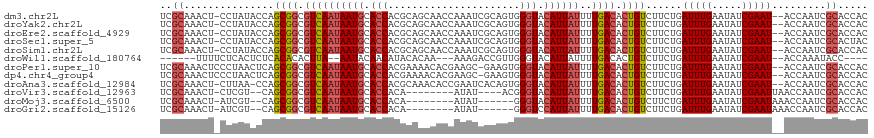
>dm3.chr2L 2784319 115 - 23011544 UCGCAAACU-CCUAUACCAGCGGCGUCAAUAAUGCACCACGCAGCAACCAAAUCGCAGUGGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAU--ACCAAUCGCACCAC ..((.....-.........((((.((((((((((.(((.(((.((.........)).)))))).))))))..)))).))))......(((((.....)))))--.......))..... ( -25.50, z-score = -1.69, R) >droYak2.chr2L 2775902 115 - 22324452 UCGCAAACU-CCUAUACCAGCGGCGUCAAUAAUGCACCACGCAGCAACCAAAUCGCAGUGGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAU--ACCAAUCGCACCAC ..((.....-.........((((.((((((((((.(((.(((.((.........)).)))))).))))))..)))).))))......(((((.....)))))--.......))..... ( -25.50, z-score = -1.69, R) >droEre2.scaffold_4929 2830635 115 - 26641161 UCGCAAACU-CCUAUACCAGCGGCGUCAAUAAUGCACCACGCAGCAACCAAAUCGCAGUGGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAU--ACCAAUCGCACCAC ..((.....-.........((((.((((((((((.(((.(((.((.........)).)))))).))))))..)))).))))......(((((.....)))))--.......))..... ( -25.50, z-score = -1.69, R) >droSec1.super_5 943789 115 - 5866729 UCGCAAACU-CCUAUACCAGCGGCGUCAAUAAUGCACCACGCAGCAACCAAAUCGCAGUGGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAU--ACCAAUCGCACUAC ..((.....-.........((((.((((((((((.(((.(((.((.........)).)))))).))))))..)))).))))......(((((.....)))))--.......))..... ( -25.50, z-score = -1.74, R) >droSim1.chr2L 2740908 115 - 22036055 UCGCAAACU-CCUAUACCAGCGGCGUCAAUAAUGCACCACGCAGCAACCAAAUCGCAGUGGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAU--ACCAAUCGCACCAC ..((.....-.........((((.((((((((((.(((.(((.((.........)).)))))).))))))..)))).))))......(((((.....)))))--.......))..... ( -25.50, z-score = -1.69, R) >droWil1.scaffold_180764 1295808 101 - 3949147 ------UUUUCUCACUCUCACACACUUA--AAUACAACAUACACAA---AAAGACCGUUGGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAU--ACCAAAUACC---- ------......................--................---.(((((.((..((........))..))...)))))...(((((.....)))))--..........---- ( -7.70, z-score = 1.14, R) >droPer1.super_10 1131958 115 - 3432795 UCGCAAACUCCCUAACUCAGCGGCGUCAAUAAUGCACCACGAAAACACGAAGC-GAAGUGGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAU--ACCAAUCGCACCAC ..((...............((((.((((((((((..((((.............-...))))...))))))..)))).))))......(((((.....)))))--.......))..... ( -21.89, z-score = -0.52, R) >dp4.chr4_group4 2122594 115 - 6586962 UCGCAAACUCCCUAACUCAGCGGCGUCAAUAAUGCACCACGAAAACACGAAGC-GAAGUGGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAU--ACCAAUCGCACCAC ..((...............((((.((((((((((..((((.............-...))))...))))))..)))).))))......(((((.....)))))--.......))..... ( -21.89, z-score = -0.52, R) >droAna3.scaffold_12984 262045 114 - 754457 UCGCAAACU-CUUAA-CCAGCGGCGUCAAUAAUGCACCACGCAAACACCGAAUCACAGUGGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAU--ACCAAUCGCACCAC ..((.....-.....-...((((.((((((((((.(((.(((...............)))))).))))))..)))).))))......(((((.....)))))--.......))..... ( -22.66, z-score = -1.28, R) >droVir3.scaffold_12963 7813202 103 + 20206255 UCGCAAACU-CUCGU--CAGCGGCGUCAAUAAUGCACCACA--------AUAU----ACGGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAUUAACCAAUCGCACCAC ..((.....-.....--..((((.((((((((((.(((.(.--------....----..)))).))))))..)))).))))....(((((((.....))))))).......))..... ( -21.90, z-score = -2.04, R) >droMoj3.scaffold_6500 11241267 101 + 32352404 UCGCAAACU-AUCGU--CAGCGGCGUCAAUAAUGCACCACA--------AUAU------GGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAUAAACCAAUCGCACCAC ..((.....-.....--..((((.((((((((((.(((...--------....------.))).))))))..)))).))))......(((((.....))))).........))..... ( -20.50, z-score = -1.36, R) >droGri2.scaffold_15126 2516650 101 + 8399593 UCGCAAACU-AUCGU--CAGCGGCGUCAAUAAUGCACCACA--------AUAU------GGGUCCAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAUAAACCAAUCGCACCAC ..((.....-.....--..((((.((((((((((.(((...--------....------.))).))))))..)))).))))......(((((.....))))).........))..... ( -20.50, z-score = -1.29, R) >consensus UCGCAAACU_CCUAUACCAGCGGCGUCAAUAAUGCACCACGCAGCAACCAAAUCGCAGUGGGUACAUUAUUUUGACACUGUCUUCUGAUUUGAAUAUCGAAU__ACCAAUCGCACCAC ..((...............((((.((((((((((.(((......................))).))))))..)))).))))......(((((.....))))).........))..... (-15.47 = -15.91 + 0.44)
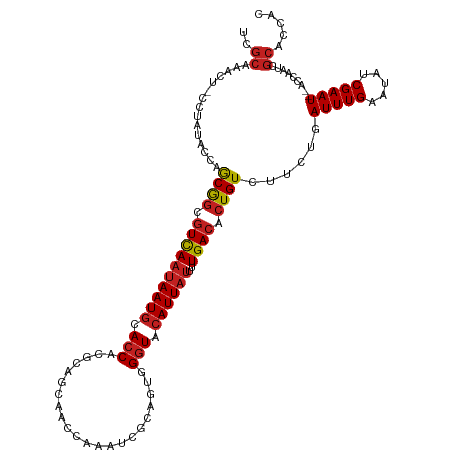
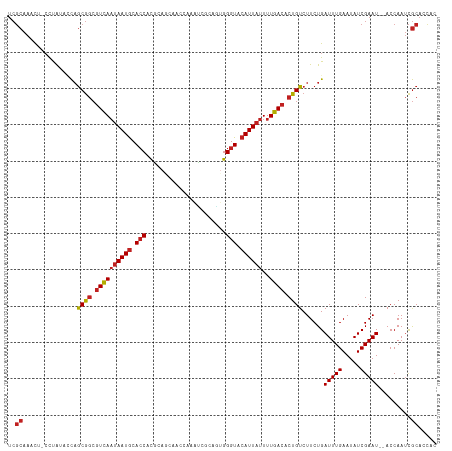
| Location | 2,784,357 – 2,784,455 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.32 |
| Shannon entropy | 0.31526 |
| G+C content | 0.42426 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -18.65 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2784357 98 + 23011544 GUGUCAAAAUAAUGUACCCACUGCGAUUUGGUUGCUGCGUGGUGCAUUAUUGACGCCGCUGGUAU-AGGAGUUUGCGAUAGUUUGUAUUAUGGAAAGAG- ((((((..(((((((((((.(.(((((...))))).).).))))))))))))))))(((......-........)))(((((....)))))........- ( -26.44, z-score = -0.87, R) >droYak2.chr2L 2775940 98 + 22324452 GUGUCAAAAUAAUGUACCCACUGCGAUUUGGUUGCUGCGUGGUGCAUUAUUGACGCCGCUGGUAU-AGGAGUUUGCGAUAGUUUGUAUUAUGGAAAGAG- ((((((..(((((((((((.(.(((((...))))).).).))))))))))))))))(((......-........)))(((((....)))))........- ( -26.44, z-score = -0.87, R) >droEre2.scaffold_4929 2830673 98 + 26641161 GUGUCAAAAUAAUGUACCCACUGCGAUUUGGUUGCUGCGUGGUGCAUUAUUGACGCCGCUGGUAU-AGGAGUUUGCGAUAGUUUGUAUUAUGGAAAGAG- ((((((..(((((((((((.(.(((((...))))).).).))))))))))))))))(((......-........)))(((((....)))))........- ( -26.44, z-score = -0.87, R) >droSec1.super_5 943827 98 + 5866729 GUGUCAAAAUAAUGUACCCACUGCGAUUUGGUUGCUGCGUGGUGCAUUAUUGACGCCGCUGGUAU-AGGAGUUUGCGAUAGUUUGUAUUAUGGAAAGAG- ((((((..(((((((((((.(.(((((...))))).).).))))))))))))))))(((......-........)))(((((....)))))........- ( -26.44, z-score = -0.87, R) >droSim1.chr2L 2740946 98 + 22036055 GUGUCAAAAUAAUGUACCCACUGCGAUUUGGUUGCUGCGUGGUGCAUUAUUGACGCCGCUGGUAU-AGGAGUUUGCGAUAGUUUGUAUUAUGGAAAGAG- ((((((..(((((((((((.(.(((((...))))).).).))))))))))))))))(((......-........)))(((((....)))))........- ( -26.44, z-score = -0.87, R) >droPer1.super_10 1131996 98 + 3432795 GUGUCAAAAUAAUGUACCCACUUCGCUUCG-UGUUUUCGUGGUGCAUUAUUGACGCCGCUGAGUUAGGGAGUUUGCGAUAGUUUGUAUUAUGGAAAGAG- ((((((..((((((((((.((...((....-.))....))))))))))))))))))(((.((.((...)).)).)))(((((....)))))........- ( -22.20, z-score = -0.04, R) >dp4.chr4_group4 2122632 98 + 6586962 GUGUCAAAAUAAUGUACCCACUUCGCUUCG-UGUUUUCGUGGUGCAUUAUUGACGCCGCUGAGUUAGGGAGUUUGCGAUAGUUUGUAUUAUGGAAAGAG- ((((((..((((((((((.((...((....-.))....))))))))))))))))))(((.((.((...)).)).)))(((((....)))))........- ( -22.20, z-score = -0.04, R) >droAna3.scaffold_12984 262083 98 + 754457 GUGUCAAAAUAAUGUACCCACUGUGAUUCGGUGUUUGCGUGGUGCAUUAUUGACGCCGCUGGUU--AAGAGUUUGCGAUAGUUUGUAUUAUGGAAGAGUU ((((((..(((((((((((((((.....))))).......)))))))))))))))).(((..((--.......(((((....))))).....))..))). ( -25.51, z-score = -1.14, R) >droVir3.scaffold_12963 7813242 84 - 20206255 GUGUCAAAAUAAUGUACCCGU------------AUAUUGUGGUGCAUUAUUGACGCCGCUGAC---GAGAGUUUGCGAUAGUUUGUAUUAUGGAAAGAG- ((((((..((((((((((((.------------....)).))))))))))))))))(((.(((---....))).)))(((((....)))))........- ( -27.90, z-score = -4.35, R) >droMoj3.scaffold_6500 11241307 82 - 32352404 GUGUCAAAAUAAUGUACCC--------------AUAUUGUGGUGCAUUAUUGACGCCGCUGAC---GAUAGUUUGCGAUAGUUUGUAUUAUGGAAAGAG- ((((((..(((((((((((--------------.....).))))))))))))))))(((.(((---....))).)))(((((....)))))........- ( -25.20, z-score = -3.49, R) >droGri2.scaffold_15126 2516690 82 - 8399593 GUGUCAAAAUAAUGGACCC--------------AUAUUGUGGUGCAUUAUUGACGCCGCUGAC---GAUAGUUUGCGAUAGUUUGUAUUAUGGAAAGAG- ((((((..((((((.((((--------------.....).))).))))))))))))(((.(((---....))).)))(((((....)))))........- ( -22.70, z-score = -2.45, R) >consensus GUGUCAAAAUAAUGUACCCACUGCGAUUUGGUUGUUGCGUGGUGCAUUAUUGACGCCGCUGGUAU_AGGAGUUUGCGAUAGUUUGUAUUAUGGAAAGAG_ ((((((..(((((((((((...................).))))))))))))))))(((...............)))(((((....)))))......... (-18.65 = -18.74 + 0.09)

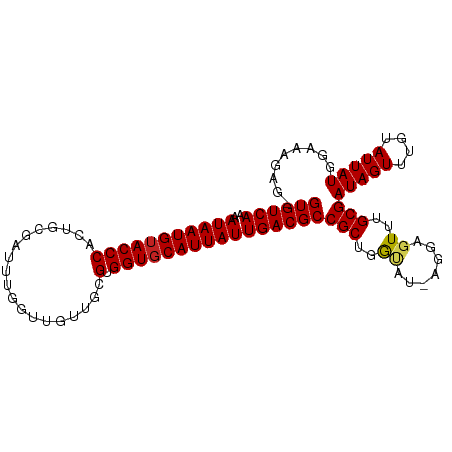
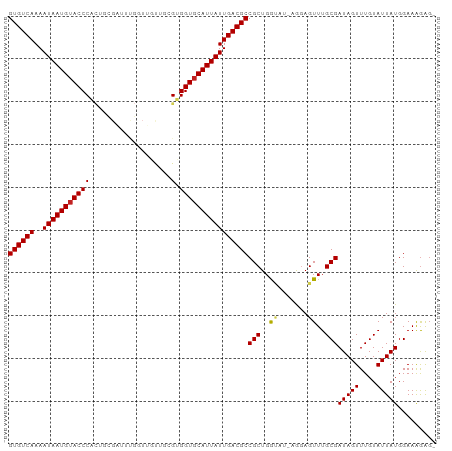
| Location | 2,784,357 – 2,784,455 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.32 |
| Shannon entropy | 0.31526 |
| G+C content | 0.42426 |
| Mean single sequence MFE | -16.97 |
| Consensus MFE | -12.01 |
| Energy contribution | -12.01 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2784357 98 - 23011544 -CUCUUUCCAUAAUACAAACUAUCGCAAACUCCU-AUACCAGCGGCGUCAAUAAUGCACCACGCAGCAACCAAAUCGCAGUGGGUACAUUAUUUUGACAC -.....................((((........-......)))).((((((((((.(((.(((.((.........)).)))))).))))))..)))).. ( -18.84, z-score = -2.02, R) >droYak2.chr2L 2775940 98 - 22324452 -CUCUUUCCAUAAUACAAACUAUCGCAAACUCCU-AUACCAGCGGCGUCAAUAAUGCACCACGCAGCAACCAAAUCGCAGUGGGUACAUUAUUUUGACAC -.....................((((........-......)))).((((((((((.(((.(((.((.........)).)))))).))))))..)))).. ( -18.84, z-score = -2.02, R) >droEre2.scaffold_4929 2830673 98 - 26641161 -CUCUUUCCAUAAUACAAACUAUCGCAAACUCCU-AUACCAGCGGCGUCAAUAAUGCACCACGCAGCAACCAAAUCGCAGUGGGUACAUUAUUUUGACAC -.....................((((........-......)))).((((((((((.(((.(((.((.........)).)))))).))))))..)))).. ( -18.84, z-score = -2.02, R) >droSec1.super_5 943827 98 - 5866729 -CUCUUUCCAUAAUACAAACUAUCGCAAACUCCU-AUACCAGCGGCGUCAAUAAUGCACCACGCAGCAACCAAAUCGCAGUGGGUACAUUAUUUUGACAC -.....................((((........-......)))).((((((((((.(((.(((.((.........)).)))))).))))))..)))).. ( -18.84, z-score = -2.02, R) >droSim1.chr2L 2740946 98 - 22036055 -CUCUUUCCAUAAUACAAACUAUCGCAAACUCCU-AUACCAGCGGCGUCAAUAAUGCACCACGCAGCAACCAAAUCGCAGUGGGUACAUUAUUUUGACAC -.....................((((........-......)))).((((((((((.(((.(((.((.........)).)))))).))))))..)))).. ( -18.84, z-score = -2.02, R) >droPer1.super_10 1131996 98 - 3432795 -CUCUUUCCAUAAUACAAACUAUCGCAAACUCCCUAACUCAGCGGCGUCAAUAAUGCACCACGAAAACA-CGAAGCGAAGUGGGUACAUUAUUUUGACAC -.....................((((...............)))).((((((((((..((((.......-.........))))...))))))..)))).. ( -15.15, z-score = -0.53, R) >dp4.chr4_group4 2122632 98 - 6586962 -CUCUUUCCAUAAUACAAACUAUCGCAAACUCCCUAACUCAGCGGCGUCAAUAAUGCACCACGAAAACA-CGAAGCGAAGUGGGUACAUUAUUUUGACAC -.....................((((...............)))).((((((((((..((((.......-.........))))...))))))..)))).. ( -15.15, z-score = -0.53, R) >droAna3.scaffold_12984 262083 98 - 754457 AACUCUUCCAUAAUACAAACUAUCGCAAACUCUU--AACCAGCGGCGUCAAUAAUGCACCACGCAAACACCGAAUCACAGUGGGUACAUUAUUUUGACAC ......................((((........--.....)))).((((((((((.(((.(((...............)))))).))))))..)))).. ( -16.08, z-score = -1.64, R) >droVir3.scaffold_12963 7813242 84 + 20206255 -CUCUUUCCAUAAUACAAACUAUCGCAAACUCUC---GUCAGCGGCGUCAAUAAUGCACCACAAUAU------------ACGGGUACAUUAUUUUGACAC -.....................((((..((....---))..)))).((((((((((.(((.(.....------------..)))).))))))..)))).. ( -14.90, z-score = -2.62, R) >droMoj3.scaffold_6500 11241307 82 + 32352404 -CUCUUUCCAUAAUACAAACUAUCGCAAACUAUC---GUCAGCGGCGUCAAUAAUGCACCACAAUAU--------------GGGUACAUUAUUUUGACAC -.....................((((..((....---))..)))).((((((((((.(((.......--------------.))).))))))..)))).. ( -15.60, z-score = -2.70, R) >droGri2.scaffold_15126 2516690 82 + 8399593 -CUCUUUCCAUAAUACAAACUAUCGCAAACUAUC---GUCAGCGGCGUCAAUAAUGCACCACAAUAU--------------GGGUCCAUUAUUUUGACAC -.....................((((..((....---))..)))).((((((((((.(((.......--------------.))).))))))..)))).. ( -15.60, z-score = -2.52, R) >consensus _CUCUUUCCAUAAUACAAACUAUCGCAAACUCCU_AUACCAGCGGCGUCAAUAAUGCACCACGCAACAACCAAAUCGCAGUGGGUACAUUAUUUUGACAC ......................((((...............)))).((((((((((.(((......................))).))))))..)))).. (-12.01 = -12.01 + -0.00)
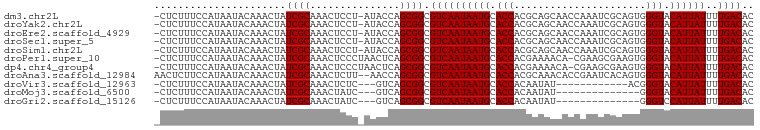

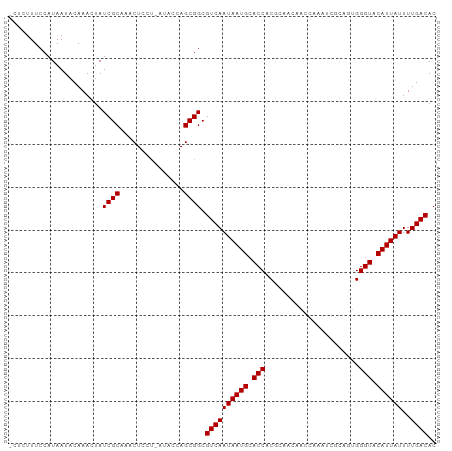
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:17 2011