| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,740,290 – 9,740,418 |
| Length | 128 |
| Max. P | 0.999334 |

| Location | 9,740,290 – 9,740,418 |
|---|---|
| Length | 128 |
| Sequences | 8 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 63.97 |
| Shannon entropy | 0.71745 |
| G+C content | 0.46591 |
| Mean single sequence MFE | -42.39 |
| Consensus MFE | -13.96 |
| Energy contribution | -12.59 |
| Covariance contribution | -1.37 |
| Combinations/Pair | 1.45 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.998851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
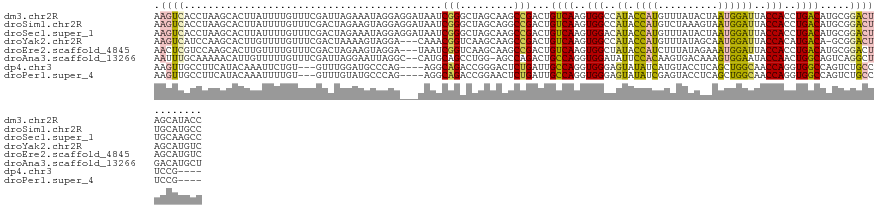
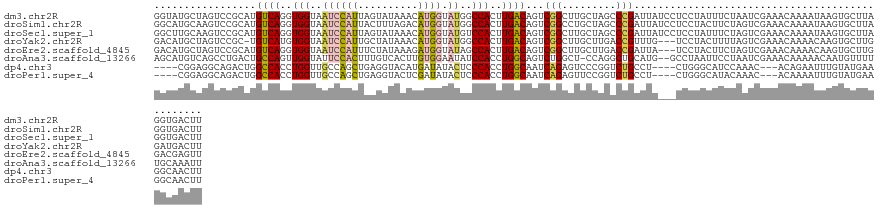
>dm3.chr2R 9740290 128 + 21146708 AAGUCACCUAAGCACUUAUUUUGUUUCGAUUAGAAAUAGGAGGAUAAUCGGGCUAGCAAGCCGACUGUCAAGUGGCCAUACCAUGUUUAUACUAAUGGAUUACCACCUGACAUGCGGACUAGCAUACC .((((.............(((((((((.....))))))))).........((((....))))))))((((.((((..((.((((..........))))))..)))).))))((((......))))... ( -34.20, z-score = -1.84, R) >droSim1.chr2R 8181856 128 + 19596830 AAGUCACCUAAGCACUUAUUUUGUUUCGACUAGAAGUAGGAGGAUAAUCGGGCUAGCAGGCCGACUGUCAAGUGGCCAUACCAUGUCUAAAGUAAUGGAUUACCACCUGACAUGCGGACUUGCAUGCC ......(((.....((((((((..........))))))))))).......(((..((((((((..(((((.((((..((.((((..........))))))..)))).)))))..))).)))))..))) ( -36.90, z-score = -1.12, R) >droSec1.super_1 7267189 128 + 14215200 AAGUCACCUAAGCACUUAUUUUGUUUCGACUAGAAAUAGGAGGAUAAUCGGGCUAGCAAGCCGACUGUCAAGUGGACAUACCAUGUUUAUACUAAUGGAUUACCACCUGACAUGCGGACUUGCAAGCC (((((......).)))).(((((((((.....))))))))).........((((.((((((((..(((((.((((.....((((..........))))....)))).)))))..))).))))).)))) ( -40.20, z-score = -3.57, R) >droYak2.chr2R 13925803 124 - 21139217 AAGUCAUCCAAGCACUUGUUUUGUUUCGACUAAAAGUAGGA---CAAACGGUCAAGCAAGCCGACUGUCAAGUGGCCAUACCAUGUUUAUAGCAAUGGAUUACCACAUGACA-GCGGACUAGCAUGUC .(((((..(((((....).))))..).))))........((---(.....)))..((.(((((.((((((.((((..((.((((((.....)).))))))..)))).)))))-)))).)).))..... ( -35.00, z-score = -2.00, R) >droEre2.scaffold_4845 6509762 125 - 22589142 AACUCGUCCAAGCACUUGUUUUGUUUCGACUAGAAGUAGGA---UAAUCGGUCAAGCAAGCCGACUGUCAAGUGGCUAUACCAUCUUUAUAGAAAUGGAUUACCACCUGACAUGCGGACUAGCAUGUC ..((.(((.(((((.......))))).))).))......((---((.(((((.......))))).))))..((((..((.((((((....)))..)))))..))))..(((((((......))))))) ( -34.90, z-score = -1.83, R) >droAna3.scaffold_13266 17041219 125 + 19884421 AAUUUGCAAAAACAUUGUUUUUGUUUCGAUUAGGAAUUAGGC--CAUGCAGCCUGG-AGCCAGACUGCCAGGUGGAUAUUCCACAAGUGACAAAGUGGAAUACCAACUGGCAGUCAGGCUGACAUGCU ((((.((((((((...))))))))...))))........(((--.((((((((((.-...).(((((((((.(((.((((((((..........))))))))))).))))))))))))))).)))))) ( -51.70, z-score = -5.27, R) >dp4.chr3 5409889 117 - 19779522 AAGUUGCCUUCAUACAAAUUCUGU---GUUUGGAUGCCCAG----AGGCAGACCGGGACUCUGAUUGCCAGGUGGGAGUAUAUCAUGUACCUCAGCUGGCAACCAGGUGGCCAGUCUGCCUCCG---- .....((.((((.(((.......)---)).)))).))...(----((((((((.((..(((((.(((((((.((((.((((.....)))))))).))))))).)))).).)).)))))))))..---- ( -51.10, z-score = -3.46, R) >droPer1.super_4 3236423 117 + 7162766 AAGUUGCCUUCAUACAAAUUUUGU---GUUUGUAUGCCCAG----AGGCAGACCGGAACUCUGAUUGCCAGGUGGGAGUAUAUCGAGUACCUCAGCUGGCAACCAGGUGGCCAGUCUGCCUCCG---- ..........(((((((((.....---)))))))))....(----((((((((.((.((.(((.(((((((.((((.((((.....)))))))).))))))).)))))..)).)))))))))..---- ( -55.10, z-score = -5.69, R) >consensus AAGUCACCUAAGCACUUAUUUUGUUUCGACUAGAAGUAGGA___UAAUCGGGCUAGCAAGCCGACUGUCAAGUGGCCAUACCAUGUUUAUAGCAAUGGAUUACCACCUGACAUGCGGACUUGCAUGCC .((((...........................................(((.........)))...((((..(((..((.((((..........))))))..)))..)))).....))))........ (-13.96 = -12.59 + -1.37)
| Location | 9,740,290 – 9,740,418 |
|---|---|
| Length | 128 |
| Sequences | 8 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 63.97 |
| Shannon entropy | 0.71745 |
| G+C content | 0.46591 |
| Mean single sequence MFE | -45.00 |
| Consensus MFE | -11.21 |
| Energy contribution | -10.28 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.91 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
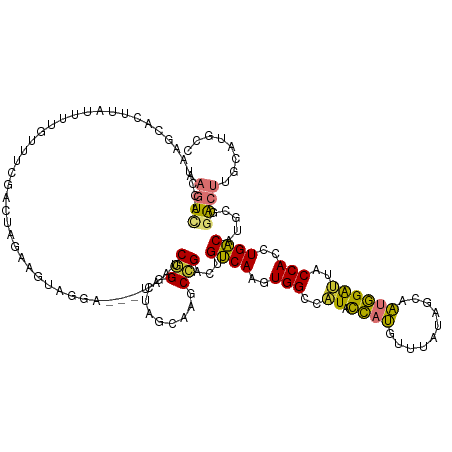
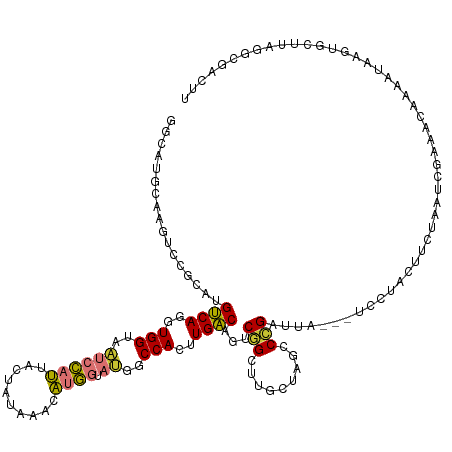
>dm3.chr2R 9740290 128 - 21146708 GGUAUGCUAGUCCGCAUGUCAGGUGGUAAUCCAUUAGUAUAAACAUGGUAUGGCCACUUGACAGUCGGCUUGCUAGCCCGAUUAUCCUCCUAUUUCUAAUCGAAACAAAAUAAGUGCUUAGGUGACUU ((((((((((((((..(((((((((((...((((..((....))))))....)))))))))))..)))...)))))).((((((............))))))...........))))).((....)). ( -40.10, z-score = -2.91, R) >droSim1.chr2R 8181856 128 - 19596830 GGCAUGCAAGUCCGCAUGUCAGGUGGUAAUCCAUUACUUUAGACAUGGUAUGGCCACUUGACAGUCGGCCUGCUAGCCCGAUUAUCCUCCUACUUCUAGUCGAAACAAAAUAAGUGCUUAGGUGACUU (((..(((.(((.((.(((((((((((...((((..........))))....))))))))))))).))).)))..)))((((((............)))))).................((....)). ( -39.10, z-score = -1.86, R) >droSec1.super_1 7267189 128 - 14215200 GGCUUGCAAGUCCGCAUGUCAGGUGGUAAUCCAUUAGUAUAAACAUGGUAUGUCCACUUGACAGUCGGCUUGCUAGCCCGAUUAUCCUCCUAUUUCUAGUCGAAACAAAAUAAGUGCUUAGGUGACUU ((((.(((((((.((.((((((((((....((((..((....)))))).....)))))))))))).))))))).))))((((((............)))))).................((....)). ( -43.40, z-score = -4.17, R) >droYak2.chr2R 13925803 124 + 21139217 GACAUGCUAGUCCGC-UGUCAUGUGGUAAUCCAUUGCUAUAAACAUGGUAUGGCCACUUGACAGUCGGCUUGCUUGACCGUUUG---UCCUACUUUUAGUCGAAACAAAACAAGUGCUUGGAUGACUU ..((.((.((((.((-(((((.((((....((((.(((((....))))))))))))).))))))).)))).)).)).....(..---((((((((................)))))...)))..)... ( -35.29, z-score = -1.61, R) >droEre2.scaffold_4845 6509762 125 + 22589142 GACAUGCUAGUCCGCAUGUCAGGUGGUAAUCCAUUUCUAUAAAGAUGGUAUAGCCACUUGACAGUCGGCUUGCUUGACCGAUUA---UCCUACUUCUAGUCGAAACAAAACAAGUGCUUGGACGAGUU .....(((.((((((((((((((((((.((((((((......)))))).)).))))))))))(((((((......).)))))).---..........................))))..)))).))). ( -40.40, z-score = -3.23, R) >droAna3.scaffold_13266 17041219 125 - 19884421 AGCAUGUCAGCCUGACUGCCAGUUGGUAUUCCACUUUGUCACUUGUGGAAUAUCCACCUGGCAGUCUGGCU-CCAGGCUGCAUG--GCCUAAUUCCUAAUCGAAACAAAAACAAUGUUUUUGCAAAUU .(((....((((.(((((((((.(((((((((((..........)))))))).))).))))))))).))))-..(((((....)--))))...........((((((.......)))))))))..... ( -50.40, z-score = -5.97, R) >dp4.chr3 5409889 117 + 19779522 ----CGGAGGCAGACUGGCCACCUGGUUGCCAGCUGAGGUACAUGAUAUACUCCCACCUGGCAAUCAGAGUCCCGGUCUGCCU----CUGGGCAUCCAAAC---ACAGAAUUUGUAUGAAGGCAACUU ----((((((((((((((..(((((((((((((.((.((.............)))).))))))))))).)).)))))))))))----)))..(((.((((.---......)))).)))(((....))) ( -53.22, z-score = -4.61, R) >droPer1.super_4 3236423 117 - 7162766 ----CGGAGGCAGACUGGCCACCUGGUUGCCAGCUGAGGUACUCGAUAUACUCCCACCUGGCAAUCAGAGUUCCGGUCUGCCU----CUGGGCAUACAAAC---ACAAAAUUUGUAUGAAGGCAACUU ----((((((((((((((..(((((((((((((.((.((.............)))).))))))))))).)).)))))))))))----)))..((((((((.---......))))))))(((....))) ( -58.12, z-score = -6.92, R) >consensus GGCAUGCAAGUCCGCAUGUCAGGUGGUAAUCCAUUACUAUAAACAUGGUAUGGCCACUUGACAGUCGGCUUGCUAGCCCGAUUA___UCCUACUUCUAAUCGAAACAAAAUAAGUGCUUAGGCGACUU .................((((..(((..((((((..........)))).))..)))..))))...(((.........)))................................................ (-11.21 = -10.28 + -0.94)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:21:04 2011