| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,693,033 – 9,693,133 |
| Length | 100 |
| Max. P | 0.966165 |

| Location | 9,693,033 – 9,693,133 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 58.56 |
| Shannon entropy | 0.70589 |
| G+C content | 0.33302 |
| Mean single sequence MFE | -21.34 |
| Consensus MFE | -7.60 |
| Energy contribution | -7.64 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9693033 100 - 21146708 ---ACCGGACGGAACAAAAGAUUGUGC------UUGAAACUUU--UGUUUUACUGCUUAGGAUAGAGCUUA--AGUUAAAGCUUUAUCACUCCUGUUCCGUUUGAAUAUCGUA ---..(((((((((((..((.....((------.((((((...--.))))))..))....((((((((((.--.....)))))))))).))..)))))))))))......... ( -34.20, z-score = -5.20, R) >droSim1.chr2R 8134418 93 - 19596830 ------------CACAAAAUAUUGUUC------UUGAAACUUUCAUGUUUUACUGCUUGGGAUAAAGCUUA--AGUUGAAGCUUUAUCACUCCCGUUCCGUUUGAAUAUCGUA ------------..........(((((------.((((((......))))))......((((((((((((.--.....))))))))....)))).........)))))..... ( -18.60, z-score = -1.78, R) >droSec1.super_1 7220197 92 - 14215200 -------------CAAAAAAAAUGUUC------UUGAAACUUUUAUGUUUUACUGCUUGGGAUAAAGCUUA--AGUUGAAGCUUUAUCACUCCCGUUCCGUUUGAAUAUCGUA -------------........((((((------.((((((......))))))......((((((((((((.--.....))))))))....)))).........)))))).... ( -19.30, z-score = -2.35, R) >droYak2.chr2R 13877786 111 + 21139217 ACUCACAGGCUUACAAAAUUAUAGUUCAAUUUUUUAAAACUUUUAAGUUUUAGGGCUUGGAAUACGAUUUA--AGCCAAAGCCCUAGCAAUUUCGGUCCCUUCGAAUAUCGUA ....((.(((((.....................((((((((....))))))))((((((((......))))--)))).)))))........(((((.....)))))....)). ( -22.90, z-score = -1.83, R) >droAna3.scaffold_13266 16994721 92 - 19884421 --------GCUUAGGAAGGAA------------GGAGAAUCCCUAUUUCUUA-UAUUUCAAACGAUUCUUGCCAGGAAAAGGUAAAUUUUUAUUAUAUUGUUUUAUUAUCUUU --------...(((((((..(------------((......))).)))))))-......(((((((..(((((.......)))))...........))))))).......... ( -11.72, z-score = 1.45, R) >consensus _________C__AAAAAAAAAUUGUUC______UUGAAACUUUUAUGUUUUACUGCUUGGGAUAAAGCUUA__AGUUAAAGCUUUAUCACUCCCGUUCCGUUUGAAUAUCGUA ..................................((((((......)))))).....(((((((((((((........))))))))....))))).................. ( -7.60 = -7.64 + 0.04)

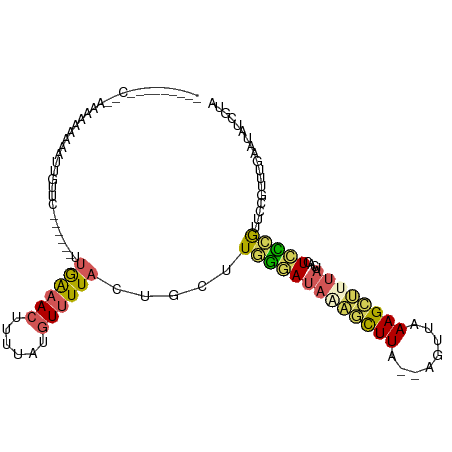
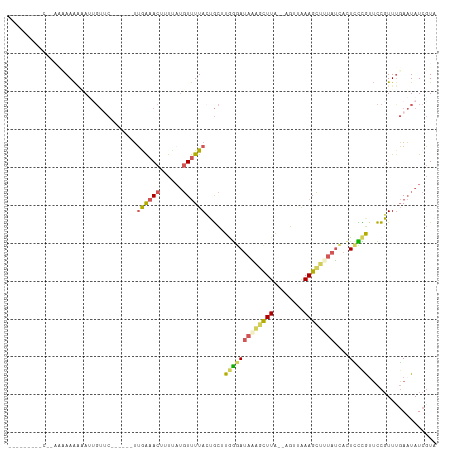
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:58 2011