
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,684,823 – 9,684,924 |
| Length | 101 |
| Max. P | 0.999101 |

| Location | 9,684,823 – 9,684,924 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 64.89 |
| Shannon entropy | 0.66970 |
| G+C content | 0.55971 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -13.75 |
| Energy contribution | -13.73 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.65 |
| SVM RNA-class probability | 0.999101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9684823 101 + 21146708 UAAAAACCAGGAGCCCAACCACCCACUGCCACA--UCCGCCCACCAAAAGGCA---GACACUCGAUAGACAAAUGGCCGCUGGCCAGUCGAGUGCACGGGCUUUUG- .......(((((((((.........(((((...--..............))))---).((((((((.......(((((...)))))))))))))...)))))))))- ( -37.94, z-score = -4.55, R) >droAna3.scaffold_13266 16987518 77 + 19884421 ----------------------------AUAGA-AUCAGGAAACCAGGGGCCACCAGACACUCGAUAGACAAAUGGCCGCUGGACAGUCGAGUGGCCGGGCUUUUG- ----------------------------.....-....(....)((((((((.........((....))....((((((((.(.....).))))))))))))))))- ( -25.30, z-score = -1.65, R) >droYak2.chr2R 13869602 103 - 21139217 UAAAAACCAGGAGCCCAGCCAGCACCACUUCCACAUCCGCCCACCAAAAGGCA---GACACUCGAUAGACAAAUGGCCGCUGGCCAGUCGAGUGCACGGGCUUUUG- .......(((((((((......................(((........))).---(.((((((((.......(((((...))))))))))))))..)))))))))- ( -35.71, z-score = -3.40, R) >droSec1.super_1 7211919 102 + 14215200 UAAAAACCAGGCGCCCAACCACC-ACUGCCACACAUCCGCCCACCAAAAGGCA---GACACUCGAUAGACAAAUGGCCGCUGGCCAGUCGAGUGCACGGGCUUUUG- .......((((.((((.......-.(((((...................))))---).((((((((.......(((((...)))))))))))))...)))).))))- ( -33.32, z-score = -2.75, R) >droSim1.chr2R 8125085 102 + 19596830 UAAAAACCAGGAGCCCAACCACC-ACUGCCACACAUCCGCCCACCAAAAGGCA---GACACUCGAUAGACAAAUGACCGCUGGCCAGUCGAGUGCACGGGCUUUUG- .......(((((((((.......-.(((((...................))))---).((((((((.(.((.........))..).))))))))...)))))))))- ( -31.51, z-score = -3.38, R) >droVir3.scaffold_12875 8344767 97 + 20611582 ------GAAGCCUUUUGGGGCCAUGCAAAAACAGAUCAUGCUAU-AAAAGGCA---AAGUUGCCGCAGCUAGACAAGCGCUAGACAGACCUAGACCCAGACCCAGGC ------...((((((((((((...(((...........)))...-....((((---....)))))).(((.....))).((((......)))).))))))...)))) ( -24.80, z-score = -0.62, R) >consensus UAAAAACCAGGAGCCCAACCACC_ACUGCCACACAUCCGCCCACCAAAAGGCA___GACACUCGAUAGACAAAUGGCCGCUGGCCAGUCGAGUGCACGGGCUUUUG_ ............((((......................(((........)))......((((((((.(.((.........))..).))))))))...))))...... (-13.75 = -13.73 + -0.02)

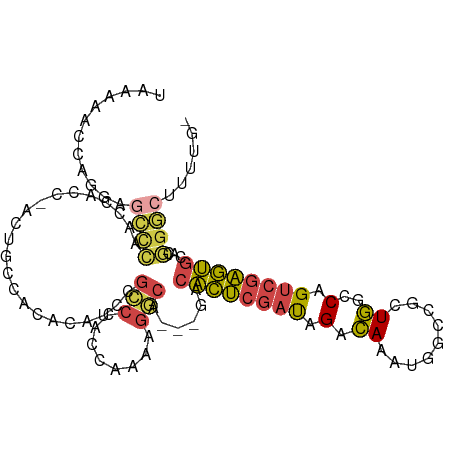
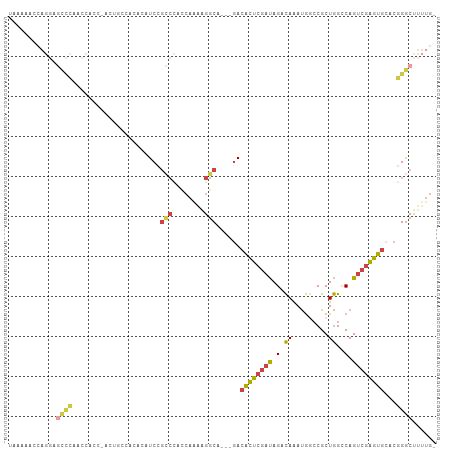
| Location | 9,684,823 – 9,684,924 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 64.89 |
| Shannon entropy | 0.66970 |
| G+C content | 0.55971 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -14.92 |
| Energy contribution | -16.98 |
| Covariance contribution | 2.07 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9684823 101 - 21146708 -CAAAAGCCCGUGCACUCGACUGGCCAGCGGCCAUUUGUCUAUCGAGUGUC---UGCCUUUUGGUGGGCGGA--UGUGGCAGUGGGUGGUUGGGCUCCUGGUUUUUA -((..((((((..(((((.((((.(((.((.(((((((.....)))))(((---..((....))..))))).--))))))))))))))..))))))..))....... ( -40.90, z-score = -1.57, R) >droAna3.scaffold_13266 16987518 77 - 19884421 -CAAAAGCCCGGCCACUCGACUGUCCAGCGGCCAUUUGUCUAUCGAGUGUCUGGUGGCCCCUGGUUUCCUGAU-UCUAU---------------------------- -((.(((((.(((((((.(((((.....))).((((((.....)))))))).)))))))...)))))..))..-.....---------------------------- ( -22.50, z-score = -0.97, R) >droYak2.chr2R 13869602 103 + 21139217 -CAAAAGCCCGUGCACUCGACUGGCCAGCGGCCAUUUGUCUAUCGAGUGUC---UGCCUUUUGGUGGGCGGAUGUGGAAGUGGUGCUGGCUGGGCUCCUGGUUUUUA -.(((((((((......)).(..(((((((.((((((.(.((((...((((---..((....))..)))))))).).)))))))))))))..)......))))))). ( -43.40, z-score = -2.15, R) >droSec1.super_1 7211919 102 - 14215200 -CAAAAGCCCGUGCACUCGACUGGCCAGCGGCCAUUUGUCUAUCGAGUGUC---UGCCUUUUGGUGGGCGGAUGUGUGGCAGU-GGUGGUUGGGCGCCUGGUUUUUA -.(((((((.(.((.((((((((.(((((.(((((((((((((((((....---.....))))))))))))))).)).))..)-)))))))))).))).))))))). ( -44.40, z-score = -2.40, R) >droSim1.chr2R 8125085 102 - 19596830 -CAAAAGCCCGUGCACUCGACUGGCCAGCGGUCAUUUGUCUAUCGAGUGUC---UGCCUUUUGGUGGGCGGAUGUGUGGCAGU-GGUGGUUGGGCUCCUGGUUUUUA -((..((((((..((((..((((.(((.((..(((((((((((((((....---.....))))))))))))))))))))))))-))))..))))))..))....... ( -37.90, z-score = -1.27, R) >droVir3.scaffold_12875 8344767 97 - 20611582 GCCUGGGUCUGGGUCUAGGUCUGUCUAGCGCUUGUCUAGCUGCGGCAACUU---UGCCUUUU-AUAGCAUGAUCUGUUUUUGCAUGGCCCCAAAAGGCUUC------ (((((((((((.(..((((((((.((((((((.....))).))((((....---))))....-.))))).))))))....).)).)))))....))))...------ ( -31.60, z-score = -0.88, R) >consensus _CAAAAGCCCGUGCACUCGACUGGCCAGCGGCCAUUUGUCUAUCGAGUGUC___UGCCUUUUGGUGGGCGGAUGUGUGGCAGU_GGUGGUUGGGCUCCUGGUUUUUA ......(((((..((((.....((((...))))((((((((((((((............))))))))))))))...........))))..)))))............ (-14.92 = -16.98 + 2.07)

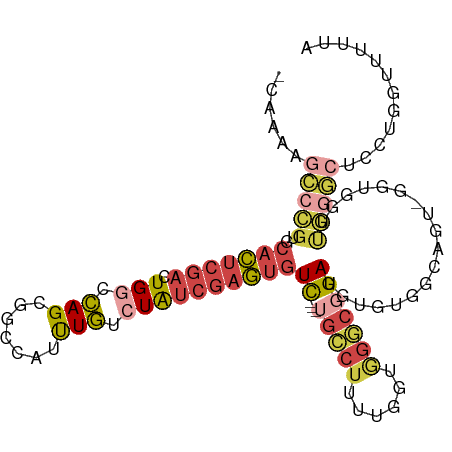
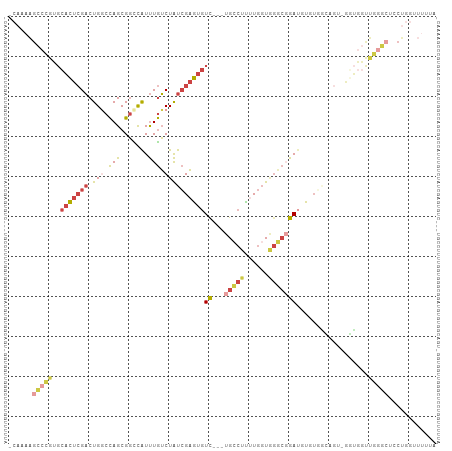
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:58 2011