
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,651,744 – 9,651,853 |
| Length | 109 |
| Max. P | 0.597985 |

| Location | 9,651,744 – 9,651,853 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.30 |
| Shannon entropy | 0.52374 |
| G+C content | 0.53424 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -13.58 |
| Energy contribution | -15.59 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
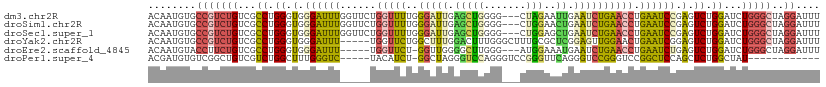
>dm3.chr2R 9651744 109 - 21146708 ACAAUGUGCCGUCUGUCGCCUGGGUGGGAUUUGGUUCUGGUUUUGGGAUUGAGCUGGGG---CUAGAAUUGAAUCUGAACCUGAAUCCGAGUCUGGAUCUGGGCUAGGAUUU ..........((((...((((((((.(((((..((((((((((..(.......)..)))---)))))))..)))))..))))..(((((....)))))..))))..)))).. ( -39.00, z-score = -2.29, R) >droSim1.chr2R 8092291 109 - 19596830 ACAAUGUGCCGUCUGUCGCCUGGGUGGGAUUUGGUUCUGGUUUUGGGAUUGAGCUGGGG---CUGGAACUGAAUCUGAACCUGAAUCCGAGUCUGGAUCUGGGCUAGGAUUU ..........((((...((((((((.(((((..((((..((((..(.......)..)))---)..))))..)))))..))))..(((((....)))))..))))..)))).. ( -39.40, z-score = -2.19, R) >droSec1.super_1 7179359 109 - 14215200 ACAAUGUGCCGUCUGUCGCCUGGGUGGGAUUUGGUUCUGGUUUUGGGAUUGAGCUGGGG---CUGGAGCUGAAUCUGAACCUGAAUCCGAGUCUGGAUCUGGGCUAGGAUUU ..........((((...(((..(((.(((((((((((.(((((..(((((.((((....---....)))).)))))))))).))).))))))))..)))..)))..)))).. ( -41.30, z-score = -2.40, R) >droYak2.chr2R 13836886 107 + 21139217 ACAAUGUGCCGUCUGUCGCCUGGGUGGGAUUU-----UGGUUCUGGCUUUGGACUUUGGGCUUUGCGCUCGGAGUUGGAACUGAAUCGGAGUCUGGAUCUGGGCUAGGAUUU ..........((((...(((..(((.((((((-----((((((.((.(((.((((((((((.....)))))))))).)))))))))))))))))..)))..)))..)))).. ( -42.20, z-score = -3.26, R) >droEre2.scaffold_4845 6422413 103 + 22589142 ACAAUGUACCUUCUGUCGCCUGGGUGGGAUUU-----UGGUUCU-GGUUGGGGCUUGGG---AUGGAAAUGAAUCUGAACCUGAAUCUGAGUCUGGAUCUGGGCUAGGAUUU ..................(((((.(.(((((.-----.(..((.-((((.(((.((.((---((........)))).))))).)))).))..)..))))).).))))).... ( -32.70, z-score = -1.67, R) >droPer1.super_4 3154413 94 - 7162766 ACGAUGUGUCGGCUGUCGUCUGGCUUUGGGUC-----UACAUCU-GGCUAGGGUCCAGGGUCCGGGUUCAGGGUCCGGGUCCGGCUCCAGCUCUGGCUAU------------ ..((((((..(((....))).(((.....)))-----))))))(-(((((((((...((..(((((..(.(....))..)))))..)).)))))))))).------------ ( -36.50, z-score = -0.20, R) >consensus ACAAUGUGCCGUCUGUCGCCUGGGUGGGAUUU_____UGGUUCUGGGAUUGAGCUGGGG___CUGGAACUGAAUCUGAACCUGAAUCCGAGUCUGGAUCUGGGCUAGGAUUU ..........((((...((((((((.((((((......(((((..(((((.(((((.......)))..)).)))))))))).......))))))..))))))))..)))).. (-13.58 = -15.59 + 2.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:53 2011