| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,644,488 – 9,644,596 |
| Length | 108 |
| Max. P | 0.572747 |

| Location | 9,644,488 – 9,644,596 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 67.17 |
| Shannon entropy | 0.70491 |
| G+C content | 0.59061 |
| Mean single sequence MFE | -42.03 |
| Consensus MFE | -15.97 |
| Energy contribution | -15.81 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.85 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.572747 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 9644488 108 - 21146708 CCUGGCUAAGCUCCUGUUGAUAGGCCAACUGGGCAGCAGGUUUCACAAAUUGGCCGGCGUAGUCUCCUUCCUUCGGCGUCCGUGGAUUCGGGCGC---UGUGGAGGAUUCU--- .((((((((...(((((((.(((.....)))..))))))).........))))))))...((..((((((...(((((((((......)))))))---)).))))))..))--- ( -46.40, z-score = -2.28, R) >droSim1.chr2R 8085090 108 - 19596830 CCUGGCUGAGCUCCUGUUGGUAGGCCAACUGGGCAGCAGGUUUCACAAAUUGGCCGGCGUAGUCUCCUUCCUUCGGCGUCCGCGGAUUCGGGCGC---UGGGGAGGAUUCU--- .(((((..((((((((((((....))))).))).))).(......)...)..)))))...((..(((((((..(((((((((......)))))))---)))))))))..))--- ( -49.60, z-score = -1.95, R) >droSec1.super_1 7172188 108 - 14215200 CCUGGCUGAGCUCCUGUUGGUAGGCCAACUGGGCAGCAGGCUUCACAAAUUGGCCGGCGUAGUCUCCUUCCUUCGGCGUCCGUGGAUUCGGGCGC---UGGGGAGGAUUCU--- ...(((((.((.((.(((((....))))).)))).((.((((.........)))).)).)))))(((((((..(((((((((......)))))))---)))))))))....--- ( -51.50, z-score = -2.37, R) >droYak2.chr2R 13829508 108 + 21139217 CCUGGCUGAGCUCCUGUUGGUAGGUCAACUGGGCAGCAGGCUUCACAAAUUGGCCGGCGUAGUCUCCUUCCGUCGGCGUCCGUGGACUCGGUCGC---UGCGGAGGAUUCU--- ((..(.(((.((((....)).)).))).)..))..((.((((.........)))).))..((..((((.(((.(((((.(((......))).)))---)))))))))..))--- ( -45.30, z-score = -1.26, R) >droEre2.scaffold_4845 6415269 108 + 22589142 CCUGGCUGAGCUCCUGUUGGAAGGUCAACUGGGCAGCAGGUUGCACAAACUGGCCGGCGUAGUCUCCAUCCUUCGGUGUCCGUGGAUUCGGGCGC---UGGGGAGGAUUCU--- .((((((..(((((((((((....))))).))).))).((((.....))))))))))...((..(((.((((.(((((((((......)))))))---)))))))))..))--- ( -44.40, z-score = -0.74, R) >droAna3.scaffold_13266 16948660 90 - 19884421 CCUGACCAAGUUCCCGUUGGUACGCCAACUGGGCGGCAGGCCGCAAAAAGUGUG------GGUUCUCCGGUUGGGGUUGUUUCUGGGGCGGAGUCU------------------ ((((.((.....((((((((....))))).))).))))))(((((.....))))------)...(((((.((.(((.....))).)).)))))...------------------ ( -34.50, z-score = -0.28, R) >dp4.chr3 5318967 108 + 19779522 CAUGACUGUAUUCCUGUUGAUAGGACAACUGGCCAGCAGGUCGCACAAAGUGGGCGGCGUAGUCCGCCUCCUGCUGUGGCCGAGGGCUCGGCAAC---UGCGGAGGACUUC--- ...(((((.....((((((.(((.....)))..))))))(((((.(.....).))))).)))))..(((((.((.((.(((((....))))).))---.))))))).....--- ( -43.20, z-score = -0.81, R) >droPer1.super_4 3146063 108 - 7162766 CAUGACUGUAUUCCUGUUGAUAGGACAACUGGCCAGCAGGUCGCACAAAGUGGGCGGCGUAGUCCGCCUCCUGCUGUGGCCGAGGGCUCGGCAAC---UGCGGAGGACUUC--- ...(((((.....((((((.(((.....)))..))))))(((((.(.....).))))).)))))..(((((.((.((.(((((....))))).))---.))))))).....--- ( -43.20, z-score = -0.81, R) >droWil1.scaffold_181009 2482239 114 + 3585778 GAUGACUUAGUUCCCGUUGAUAGGACAACUGGGCGGCUGGUCUCACAAACUGUGCCGAAUAGUCACUCUCUUUAGGCGGUUGCGAAUUAUGUUGCAAUUGUGGUGGAUGAUUAU ((.((((.(((((((((((......)))).))).))))))))))..............(((((((..(.((....((((((((((......)))))))))))).)..))))))) ( -32.40, z-score = -0.69, R) >droMoj3.scaffold_6496 7927409 105 + 26866924 ---GACUCAACUCCUGCUGAGGGCUCAACUGGCCAGCGGGUCUGAUGAAGGGACUAUCGUAGGCGAUUUCCUUGGGCUGCUUGGCAAUGAUGCGC---UGCGGGGGAUUGU--- ---....((((((((((..((.(((((....((((((((.((.....((((((..((((....)))))))))))).)))).))))..))).)).)---)))))))).))).--- ( -39.80, z-score = -1.16, R) >droVir3.scaffold_12875 8296186 105 - 20611582 ---GACUCAACUCCUGUUGGUAGCUCAACUGGGCGGCGGGUCUUACAAACGGACCCUCGUAGGCAACUUCCUUGGGCGGAUUGUAGAGUGUGCGU---GCUGGUGGAUUGU--- ---..........(..(..(((((((....)))).(((.(..((((((.(.(.(((....((....)).....)))).).))))))..).))).)---))..)..).....--- ( -32.00, z-score = 0.86, R) >consensus CCUGACUGAGCUCCUGUUGGUAGGCCAACUGGGCAGCAGGUCUCACAAAGUGGCCGGCGUAGUCUCCUUCCUUCGGCGGCCGUGGAUUCGGGCGC___UGCGGAGGAUUCU___ ...((((.....(((((((.(((.....)))..)))))))...((.....))........))))..((.((....(((((((......)))))))......)).))........ (-15.97 = -15.81 + -0.16)

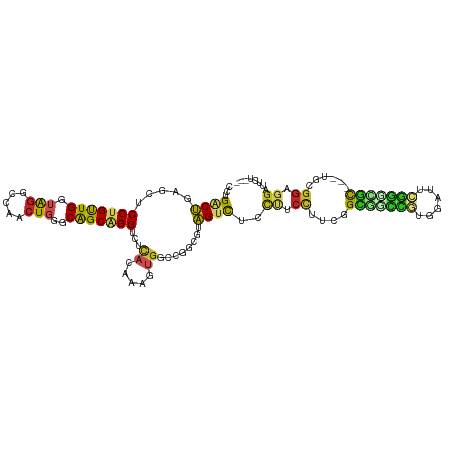

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:51 2011