| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,644,154 – 9,644,263 |
| Length | 109 |
| Max. P | 0.883420 |

| Location | 9,644,154 – 9,644,263 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 65.56 |
| Shannon entropy | 0.69468 |
| G+C content | 0.51680 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -11.35 |
| Energy contribution | -10.16 |
| Covariance contribution | -1.18 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
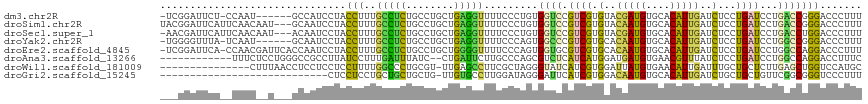
>dm3.chr2R 9644154 109 - 21146708 -UCGGAUUCU-CCAAU------GCCAUCCUACCUUUGCCUCUGCCUGCUGAGGUUUUCCCUGUGGUCCGUCGUGUACGAUGUGCACAUUGAUCUCCUGAUCCUGACCGGGACCCUUU -..((((.(.-.....------)..)))).......(((((........))))).........((((((((((((((...))))))...((((....))))..)))..))))).... ( -28.70, z-score = -1.36, R) >droSim1.chr2R 8084751 114 - 19596830 UACGGAUUCAUUCAACAAU---GCAAUCCUACCUUUGCCUCUGCCUGCUGAGGUUUUCCCUGUGGUCCGUCGUGUACAAUGUGCACAUUGAUCUCCUGAUCCUGACCGGGACCCUUU ...(((((((((....)))---).))))).......(((((........))))).........((((((((((((((...))))))...((((....))))..)))..))))).... ( -33.20, z-score = -2.72, R) >droSec1.super_1 7171850 113 - 14215200 -AACGAUUCAUUCAACAAU---ACAAUCCUACCUUUGCCUCUGCCUGCUGAGGUUUUCCCUGUGGUCCGUCGUGUACGAUGUGCACAUUGAUCUCCUGAUCCUGACCUGGACCCUUU -..................---(((.....(((((.((........)).)))))......)))((((((((((((((...))))))...((((....))))..)))..))))).... ( -27.60, z-score = -2.46, R) >droYak2.chr2R 13829174 109 + 21139217 -UGGGGUUUA-UCAAU------GCAAUCCUACCUUUGCCUCUGCCUGCUGAGGUUUUCCCAGUGGCCCGUCGUGCACAAUGUGCACAUUGAUCUCCUGAUCCUGGCCGGGACCCUUU -.((((((..-.....------((((........)))).....((.((((.((....))))))((((....((((((...))))))...((((....))))..)))))))))))).. ( -36.90, z-score = -1.86, R) >droEre2.scaffold_4845 6414929 115 + 22589142 -UCGGAUUCA-CCAACGAUUCACCAAUCCUACCUUUGCCUCUGCCUGCUGGGGUUUUCCCAGUGGUGCGUCGUGCACAAUGUGCACAUUGAUCUCCUGAUCCUGGCCAGGACCCUUU -..(((((..-.............)))))..(((..(((...(((..(((((.....)))))..).))...((((((...))))))...((((....))))..))).)))....... ( -36.06, z-score = -1.72, R) >droAna3.scaffold_13266 16948391 103 - 19884421 ------------UUUCUCCUGGGCCGCCUUAUCCUUUGAUUUAUC--CUGAUUCUUGCCCAGCGUCUCAUCAUGGAUGAUGUGAACGUUUAUCUCCUGAUCCUGGCCAGGACCUUUC ------------....((((((.((((.((((((..((((....(--(((.........))).)....)))).)))))).))).......(((....)))...).))))))...... ( -24.70, z-score = -0.91, R) >droWil1.scaffold_181009 2481955 101 + 3585778 ---------------CUUUAACCUCCUCCUCCUUUUGGCCCUGCGU-UUGAGCCUUCGCUAGGGUAUCAUCGUGGAUUAUGUGAACAUUGAUUUGCUGCUCUUGAGCUGGUCCAUGC ---------------.....(((..(((.........((((((((.-.........))).)))))......(..(((((((....)).)))))..).......)))..)))...... ( -24.00, z-score = -0.24, R) >droGri2.scaffold_15245 1746020 88 - 18325388 ----------------------------CUCCUCCUGCUGCUGCUG-UUGUGCCUUGGAUAGGGAUUCAUCGUGGACAAUGUGCACAUUGAUCUGCUGCUGUUCGGCGGGUCCCUUU ----------------------------....(((.((.((....)-)...))...))).((((((((...(..(((((((....))))).))..).(((....))))))))))).. ( -27.90, z-score = -0.84, R) >consensus ___GGAUUCA__CAAC__U___GCAAUCCUACCUUUGCCUCUGCCUGCUGAGGUUUUCCCAGUGGUCCGUCGUGGACAAUGUGCACAUUGAUCUCCUGAUCCUGACCGGGACCCUUU ...............................(((..(((((........))))).........((((.((((.(..(((((....)))))..)...))))...)))))))....... (-11.35 = -10.16 + -1.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:50 2011