| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,614,976 – 9,615,086 |
| Length | 110 |
| Max. P | 0.585815 |

| Location | 9,614,976 – 9,615,086 |
|---|---|
| Length | 110 |
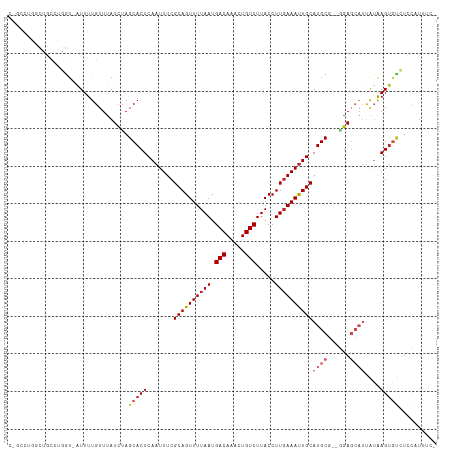
| Sequences | 11 |
| Columns | 114 |
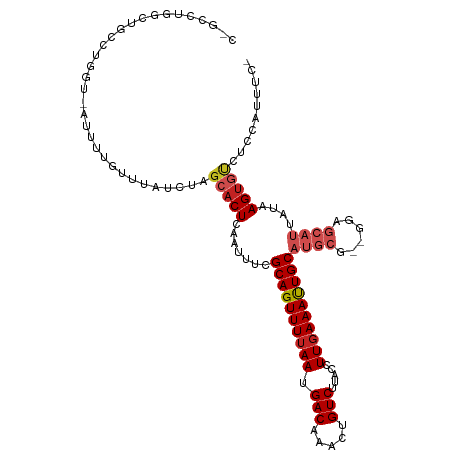
| Reading direction | reverse |
| Mean pairwise identity | 81.90 |
| Shannon entropy | 0.38972 |
| G+C content | 0.40161 |
| Mean single sequence MFE | -26.29 |
| Consensus MFE | -14.83 |
| Energy contribution | -15.80 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9614976 110 - 21146708 CUGCCUGGCUGCCUGGU-AUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCG--GGAGCAUUAUAAGUGUCUCCAUUUC- .(((.((..(((..(((-(.......))))..)))..)).....((((((((((.(((.....))).....))))))))))..)))--(((((((.....))).)))).....- ( -27.30, z-score = -1.57, R) >droSim1.chr2R 8062451 110 - 19596830 CUGCCUGGCUGCCUGGU-AUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCG--GGAGCAUUAUAAGUGUCUCCAUUUC- .(((.((..(((..(((-(.......))))..)))..)).....((((((((((.(((.....))).....))))))))))..)))--(((((((.....))).)))).....- ( -27.30, z-score = -1.57, R) >droSec1.super_1 7150524 110 - 14215200 CUGCCUGGCUGCCUGGU-AUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCG--GGAGCAUUAUAAGUGUCUCCAUUUC- .(((.((..(((..(((-(.......))))..)))..)).....((((((((((.(((.....))).....))))))))))..)))--(((((((.....))).)))).....- ( -27.30, z-score = -1.57, R) >droYak2.chr2R 13807301 110 + 21139217 CUGCCUGGCUGCCUGGU-AUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCG--GGAGCAUUAUAAGUGUCUCCAUUUC- .(((.((..(((..(((-(.......))))..)))..)).....((((((((((.(((.....))).....))))))))))..)))--(((((((.....))).)))).....- ( -27.30, z-score = -1.57, R) >droEre2.scaffold_4845 6392333 110 + 22589142 CUGCCUGGCUGCCUGGU-AUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCG--GGAGCAUUAUAAGUGUCUCCAUUUC- .(((.((..(((..(((-(.......))))..)))..)).....((((((((((.(((.....))).....))))))))))..)))--(((((((.....))).)))).....- ( -27.30, z-score = -1.57, R) >droAna3.scaffold_13266 16926308 108 - 19884421 --CGGGGACCCCUUUAC-AUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCG--GGAGCAUUAUAAGUGUCUCCAUUUC- --.((((((..(((...-((..(((((.....(((..((((((((((((((.......)))))))........)))))))..))).--.)))))..))))).)))))).....- ( -26.50, z-score = -2.17, R) >dp4.chr3 5299285 98 + 19779522 ------CUCCAUUUGGU-AUUUUGUUUAUCCAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCG--GGA----CGGGAGUCCCUUCCUC--- ------.......((((-(.......)).)))(((..((((((((((((((.......)))))))........)))))))..)))(--(((----(....)))))......--- ( -25.30, z-score = -2.18, R) >droPer1.super_4 3126336 98 - 7162766 ------CUCCAUUUGGU-AUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCG--GGA----CGGGAGUCCCUUCCUC--- ------........(((-(.......))))..(((..((((((((((((((.......)))))))........)))))))..)))(--(((----(....)))))......--- ( -25.10, z-score = -2.31, R) >droWil1.scaffold_181009 2447129 113 + 3585778 CAUCCUCCUUUUGGAGUGGUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAAAUGUCUUACCUUGAAAUUGCAUGCGAGAGAGCAUUAUAAGUGU-UGUGUGUCC .(((((((....)))).)))......(((.(((((((.......((((((((((.(((.....))).....))))))))))((((......))))....)))))-)).)))... ( -27.90, z-score = -1.80, R) >droVir3.scaffold_12875 8261627 109 - 20611582 ---ACUCCCGACAGCGU-AUUUUGUUUAUACAGCACUCAAUUUCGCAGUUUUAAUGACAAAAUGUCUUACCUUCAAAUUGCAUGCGAUAGAGCAUUAUAAGAGAGUAUGUGUG- ---......(((((...-...)))))((((((..((((......((((((((((.(((.....)))))).....)))))))((((......)))).......)))).))))))- ( -21.10, z-score = -0.50, R) >droMoj3.scaffold_6496 7896203 108 + 26866924 ---GCUCCCG-CUGCGU-AUUUUGUUUAUACAGCACUCAAUUUCGCAGUUUUAAUGACAAAAUGUCUUACCUUGAAACUGCAUGCGAGAUAGCAUUAUAAGUGUGUAUGUGUU- ---.....((-((((((-((.......)))).(((((.......((((((((((.(((.....))).....))))))))))((((......))))....)))))))).)))..- ( -26.80, z-score = -1.56, R) >consensus C_GCCUGGCUGCCUGGU_AUUUUGUUUAUCUAGCACUCAAUUUCGCAGUUUUAAUGACAAACUGUCUUACCUUGAAAUUGCAUGCG__GGAGCAUUAUAAGUGUCUCCAUUUC_ ................................(((((.......((((((((((.(((.....))).....))))))))))((((......))))....))))).......... (-14.83 = -15.80 + 0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:47 2011