| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,589,033 – 9,589,130 |
| Length | 97 |
| Max. P | 0.500000 |

| Location | 9,589,033 – 9,589,130 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.27 |
| Shannon entropy | 0.43960 |
| G+C content | 0.41815 |
| Mean single sequence MFE | -16.95 |
| Consensus MFE | -9.71 |
| Energy contribution | -9.61 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2R 9589033 97 + 21146708 AGCAACGGCA--GCAAAAAUUGCUCUUAAUUGGCGGCAGCCAUUAAAUGCAAUUUAUCAAACAGACGACAGAGACA---GCCACACACAAAGCACAGCAGCA------------ .((...(((.--....(((((((..(((((.(((....))))))))..))))))).((.....))...........---))).........((...)).)).------------ ( -21.40, z-score = -1.62, R) >droSim1.chr2R 8036174 94 + 19596830 AGCAACGGCA--GCAAAAAUUGCUCUUAAUUGGCGGCAGCCAUUAAAUGCAAUUUAUCAAACAGACGACAGAGACA---GGCACACAAAGCACACAACA--------------- .((....)).--((..(((((((..(((((.(((....))))))))..))))))).....................---(.....)...))........--------------- ( -17.00, z-score = -0.94, R) >droSec1.super_1 7125024 94 + 14215200 AGCAACGGCA--GCAAAAAUUGCUCUUAAUUGGCGGCAGCCAUUAAAUGCAAUUUAUCAAACAGACGACAGAGACA---GCCACACAAAGCACACAACA--------------- ......(((.--....(((((((..(((((.(((....))))))))..))))))).((.....))...........---))).................--------------- ( -19.20, z-score = -1.90, R) >droYak2.chr2R 13781018 94 - 21139217 AGCAACGGCA--GCAAAAAUUGCUCUUAAUUGGCGGCAGCCAUUAAAUGCAAUUUAUCAAACAGACGACAGAGACA---GCCACACAAAGCACACAACA--------------- ......(((.--....(((((((..(((((.(((....))))))))..))))))).((.....))...........---))).................--------------- ( -19.20, z-score = -1.90, R) >droEre2.scaffold_4845 6367463 94 - 22589142 AGCAACGGCA--GCAAAAAUUGCUCUUAAUUGGCGGCAGCCAUUAAAUGCAAUUUAUCAAACAGACGACAGAGACA---GCCACACAAAGCACACAACA--------------- ......(((.--....(((((((..(((((.(((....))))))))..))))))).((.....))...........---))).................--------------- ( -19.20, z-score = -1.90, R) >droAna3.scaffold_13266 16899170 92 + 19884421 AGCAACGGCA--GCAAAAAUUGCUCUUAAUUGGCGGCAGCCAUUAAAUGCAAUUUAUCAAACAGACG--AGAGACA---GCCGCACAAAGCACACAAGA--------------- .((..((((.--....(((((((..(((((.(((....))))))))..)))))))..........(.--...)...---))))......))........--------------- ( -22.20, z-score = -2.04, R) >dp4.chr3 5278145 82 - 19779522 GACAGCGGCAGAGCAAAAAUUGCUCUUAAUUGGCGGCAGCCAUUAAAUGCAAUUUAUCAAACAGACGACAGACAA------CACACAC-------------------------- ....((......))..(((((((..(((((.(((....))))))))..)))))))....................------.......-------------------------- ( -16.70, z-score = -2.19, R) >droPer1.super_4 3103723 83 + 7162766 GACAGCGGCAGAGCAAAAAUUGCUCUUAAUUGGCGGCAGCCAUUAAAUGCAAUUUAUCAAACAGACGACAGACAA------CACACACA------------------------- ....((......))..(((((((..(((((.(((....))))))))..)))))))....................------........------------------------- ( -16.70, z-score = -2.05, R) >droWil1.scaffold_181009 2411205 107 - 3585778 -----UAGUA--GCAAAAAUUGCUCUUAAUUGGCGGCAGCCAUUAAAUGCAAUUUAUCAAAGAGACGACAGACAGAUAGACCAGGCAACAGCAACAAUAACAAGAAAAAGGACU -----..((.--((..(((((((..(((((.(((....))))))))..)))))))............................(....).)).))................... ( -19.50, z-score = -1.99, R) >droMoj3.scaffold_6496 7852583 89 - 26866924 ------AGCA--GCAAAAAU-GUUCUUAAUUGGCGUCAACCAUUAAAUGCAAUUUAUCAAACAGACGACACAGGCA----CAGCAUAAAAUACACAUGCACA------------ ------.(((--(((....)-))).....(((.((((......(((((...))))).......))))...))))).----..((((.........))))...------------ ( -9.12, z-score = 1.32, R) >droVir3.scaffold_12875 8226289 87 + 20611582 ------AGCA--GCAAAAAU-GUUCUUAAUUGGCGUCAGCCAUUAAAUGCAAUUUAUCAAACAGACGACA--GGCA----CAGCAUAAAAUAUACACACACA------------ ------....--((......-....(((((.(((....)))))))).(((...(..((.....))..)..--.)))----..))..................------------ ( -11.60, z-score = -0.24, R) >droGri2.scaffold_15245 1686226 87 + 18325388 ------AGCA--GCAAAAAU-GUUCUUAAUUGGCGUCAGCCAUUAAAUGCAAUUUAUCAAACAGACGACA--GGCA----CAGCAUAAAAUACCCAUACACA------------ ------....--((......-....(((((.(((....)))))))).(((...(..((.....))..)..--.)))----..))..................------------ ( -11.60, z-score = -0.04, R) >consensus AGCAACGGCA__GCAAAAAUUGCUCUUAAUUGGCGGCAGCCAUUAAAUGCAAUUUAUCAAACAGACGACAGAGACA___GCCACACAAAACACACAACA_______________ ................((((.((..(((((.(((....))))))))..)).))))........................................................... ( -9.71 = -9.61 + -0.10)

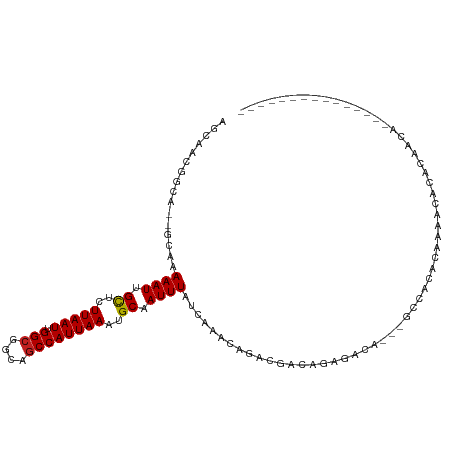

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:46 2011