
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,585,811 – 9,585,905 |
| Length | 94 |
| Max. P | 0.586995 |

| Location | 9,585,811 – 9,585,905 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Shannon entropy | 0.41012 |
| G+C content | 0.53315 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -18.32 |
| Energy contribution | -20.30 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
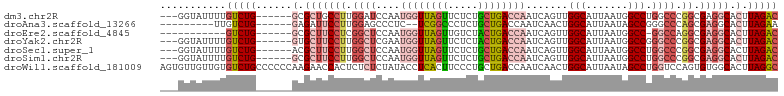
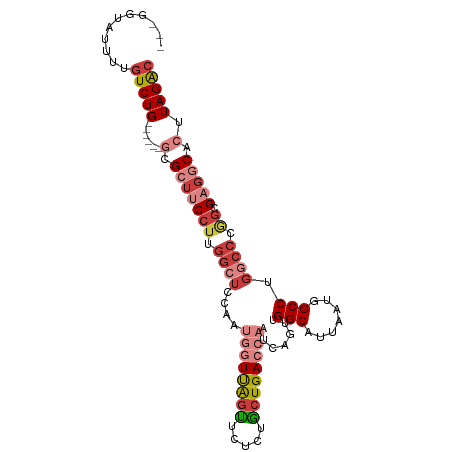
>dm3.chr2R 9585811 94 - 21146708 ---GGUAUUUUGUCUG------GCGCUGCCUUGGAUCCAAUGGUUAGUUCUCUGCUGACCAAUCAGUUGGCAUUAAUGGCCUGGCCCGGCGAGGCACUUAGAC ---(((.(((((((.(------((.(.(((...(((((((((((((((.....)))))))).....)))).)))...)))..)))).))))))).)))..... ( -32.30, z-score = -1.03, R) >droAna3.scaffold_13266 16896199 86 - 19884421 ---------UUGUCUG------GAGAUUCCUUGGAGCCCUC--UCGGCCCUCUGCUGACCAAUCAACUGGCAUUAAUAGCCGGGCCCAGCGAGGCACUUAGAA ---------.((((((------(.....))((((.((((..--(((((.....)))))..........(((.......)))))))))))..)))))....... ( -26.50, z-score = -0.05, R) >droEre2.scaffold_4845 6364459 85 + 22589142 -----------GUCUG------GCGCUUCCUCGGCUCCAAUGGUUAGUUGUCUACUGACCAAUCAGUUGGCAUUAAUGGCC-GGCCAGGCGAGGCACUUAGAC -----------(((((------(.((((((..((((....((((((((.....)))))))).......(((.......)))-))))..).))))).).))))) ( -33.50, z-score = -2.56, R) >droYak2.chr2R 13777756 94 + 21139217 ---GGUAUUUUGUCUG------GUGCUUCCUUGGCUCGAAUGGUUAGUUCUCUACUGACCAAUCAGUUGGCAUUAAUGGCCGGGCCCGGCGAGGCACUUAGAC ---........(((((------(((((((((.(((((((.((((((((.....)))))))).))....(((.......)))))))).)).))))))).))))) ( -43.00, z-score = -4.91, R) >droSec1.super_1 7121909 94 - 14215200 ---GGUAUUUUGUCUG------ACGCUUCCUUGGCUCCAAUGGUUAGUUCUCUGCUGACCAAUCAGUUGGCAUUAAUGGCCUGGCCCGGCGAGGCACUUAGAC ---........(((((------(.(((((((.((((....((((((((.....)))))))).......(((.......))).)))).)).)))))..)))))) ( -32.70, z-score = -1.87, R) >droSim1.chr2R 8031875 94 - 19596830 ---GGUAUUUUGUCUG------GCGCUUCCUUGGCUCCAAUGGUUAGUUCUCUGCUGACCAAUCAGUUGGCAUUAAUGGCCUGGCCCGGCGAGGCACUUAGAC ---........(((((------(.(((((((.((((....((((((((.....)))))))).......(((.......))).)))).)).))))).).))))) ( -33.10, z-score = -1.46, R) >droWil1.scaffold_181009 2406652 103 + 3585778 AGUGUUGUUGUGUCUGCCCCCCAAGAACCACUCUCUCUAUACCUCACUUCCCUGCUGACCAAUCAACUGGCAUUAAUAGCCUGGUCCAGUGUGGCACUUAGGC (((((..(.((((((........)))..)))......................((((((((.......(((.......))))))).)))))..)))))..... ( -19.21, z-score = 1.42, R) >consensus ___GGUAUUUUGUCUG______GCGCUUCCUUGGCUCCAAUGGUUAGUUCUCUGCUGACCAAUCAGUUGGCAUUAAUGGCCUGGCCCGGCGAGGCACUUAGAC ...........(((((........(((((((.((((....((((((((.....)))))))).......(((.......))).)))).)).)))))...))))) (-18.32 = -20.30 + 1.98)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:45 2011