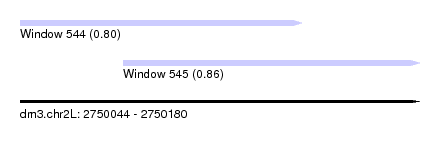
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,750,044 – 2,750,180 |
| Length | 136 |
| Max. P | 0.855089 |

| Location | 2,750,044 – 2,750,140 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 70.84 |
| Shannon entropy | 0.49323 |
| G+C content | 0.59498 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
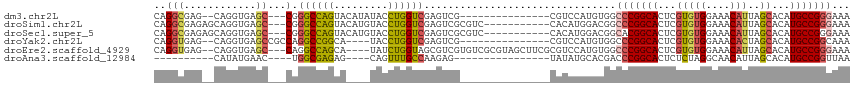
>dm3.chr2L 2750044 96 + 23011544 CAGGCGAG--CAGGUGAGC---CGGGCCAGUACAUAUACCUGGUCGAGUCG---------------CGUCCAUGUGGCCCGGCACUCGUGUGGAAACAUUAGCACAUGCCGGGAAA ....((.(--((.(((.((---((((((((((....))).(((.((.....---------------)).)))..))))))))).((.((((....)))).))))).)))))..... ( -38.80, z-score = -1.53, R) >droSim1.chr2L 2712294 102 + 22036055 CAGGCGAGAGCAGGUGAGC---CGGGCCAGUACAUGUACCUGGUCGAGUCGCGUC-----------CACAUGGACGGCCCGGCACUCGUGUGGAAACAUUAGCACAUGCCGGGAAA ..((((.(.((......((---((((((.((.(((((....((.((.....)).)-----------)))))).))))))))))......(((....)))..)).).))))...... ( -41.80, z-score = -1.62, R) >droSec1.super_5 916175 102 + 5866729 CAGGCGAGAGCAGGUGAGC---CGGGCCAGUACAUGUACCUGGUCGAGUCGCGUC-----------CACAUGGACGGCACGGCACUCGUGUGGAAACAUUAGCACAUGCCGGGAAA ..((((.(.((......((---((((((((((....)).)))))).....(((((-----------(....)))).)).))))......(((....)))..)).).))))...... ( -37.40, z-score = -0.71, R) >droYak2.chr2L 2745578 95 + 22324452 CAGGUGAG--CAGGUGAGCCGCCAGGCCGGCA----UACCUGGUCGAGUCG---------------CGUCCAUGUGGCCCGGCACUCGUGUGGAAACACUAGCACAUGCCGGCAAA ..((((.(--(......))))))..(((((((----(.....((((.((((---------------((....)))))).)))).((.((((....)))).))...))))))))... ( -42.20, z-score = -1.43, R) >droEre2.scaffold_4929 2802874 107 + 26641161 CAGGUGAG--CAGGUGAGC---CAGGCCAGCA----UAUCUGGUAGCGUCGUGUCGCGUAGCUUCGCGUCCAUGUGGCCCGGCACUCGUGUGGAAACAUUAGCACAUGCCGGGAAA ((.(((..--(..((((((---...(((((..----...))))).(((......)))...)).)))))..))).)).(((((((((.((((....)))).))....)))))))... ( -38.60, z-score = -0.02, R) >droAna3.scaffold_12984 218917 82 + 754457 ----------CAUAUGAAC----UGGCGAGAG----CAGUUUGCCAAGAG----------------UAUAUGCACGACCCGGCACUCUCUAGGCAACAUUAGCACAUGCCGGUUAA ----------......(((----(((((.(.(----(((((((((.((((----------------....(((.((...)))))..)))).))))).))).)).).)))))))).. ( -28.50, z-score = -3.07, R) >consensus CAGGCGAG__CAGGUGAGC___CGGGCCAGUA____UACCUGGUCGAGUCG_______________CAUACAUACGGCCCGGCACUCGUGUGGAAACAUUAGCACAUGCCGGGAAA ........................((((((.........))))))................................(((((((...(((((....)))..))...)))))))... (-21.04 = -21.27 + 0.23)
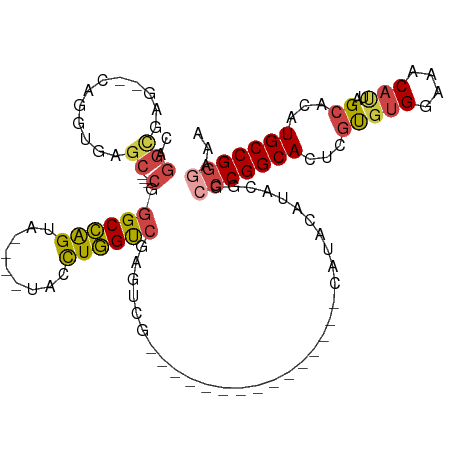

| Location | 2,750,079 – 2,750,180 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.50 |
| Shannon entropy | 0.30271 |
| G+C content | 0.58873 |
| Mean single sequence MFE | -40.18 |
| Consensus MFE | -27.00 |
| Energy contribution | -27.80 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2750079 101 + 23011544 ---------------UGGUCGAGUCGCGUCCAUGUGG----CCCGGCACUCGUGUGGAAACAUUAGCACAUGCCGGGAAAACGAGACCCGACUCCGAAAGUGUCCAAUGCGACGGCGACG ---------------..((((.((((((((..(((..----(((((((((.((((....)))).))....)))))))...))).)))..(((..(....).)))....)))))..)))). ( -42.10, z-score = -2.38, R) >droSim1.chr2L 2712331 105 + 22036055 ---------------UGGUCGAGUCGCGUCCACAUGGACGGCCCGGCACUCGUGUGGAAACAUUAGCACAUGCCGGGAAAACGAGACCCGACUGCGAAAGUGUCCAAUGCGACGGCGAUG ---------------..((((.(((((((((....))))..(((((((((.((((....)))).))....)))))))............(((.((....)))))....)))))..)))). ( -43.40, z-score = -2.39, R) >droSec1.super_5 916212 105 + 5866729 ---------------UGGUCGAGUCGCGUCCACAUGGACGGCACGGCACUCGUGUGGAAACAUUAGCACAUGCCGGGAAAACGAGACCCGACUGCGAAAGUGUCCAAUGCGACGGCGAUG ---------------..((((.(((((((((....))))(((((((((((.((((....)))).))....))))(((.........)))..........)))))....)))))..)))). ( -40.10, z-score = -1.57, R) >droYak2.chr2L 2745612 90 + 22324452 ---------------UGGUCGAGUCGCGUCCAUGUGG----CCCGGCACUCGUGUGGAAACACUAGCACAUGCCGGCAAAACGAGACUCGACUGCGAAAGUGUCCAAUG----------- ---------------.(((((((((.(((((....))----.((((((((.((((....)))).))....))))))....))).)))))))))((....))........----------- ( -37.00, z-score = -2.81, R) >droEre2.scaffold_4929 2802905 105 + 26641161 UGGUAGCGUCGUGUCGCGUAGCUUCGCGUCCAUGUGG----CCCGGCACUCGUGUGGAAACAUUAGCACAUGCCGGGAAAACGAGACUCGACUCGGAAAGUGUCCAAUG----------- (((..((.(((.((((((......)))(((..(((..----(((((((((.((((....)))).))....)))))))...))).)))..))).)))...))..)))...----------- ( -38.30, z-score = -1.18, R) >consensus _______________UGGUCGAGUCGCGUCCAUGUGG____CCCGGCACUCGUGUGGAAACAUUAGCACAUGCCGGGAAAACGAGACCCGACUGCGAAAGUGUCCAAUGCGACGGCGA_G ...............((((((.(((.(((((....))....(((((((...(((((....)))..))...)))))))...))).))).)))))).......................... (-27.00 = -27.80 + 0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:13 2011