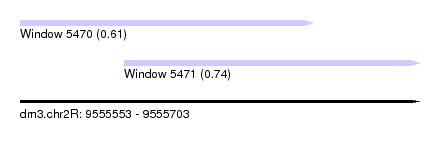
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,555,553 – 9,555,703 |
| Length | 150 |
| Max. P | 0.735584 |

| Location | 9,555,553 – 9,555,663 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.55 |
| Shannon entropy | 0.44986 |
| G+C content | 0.49144 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -20.47 |
| Energy contribution | -19.73 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
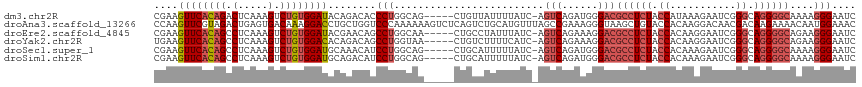
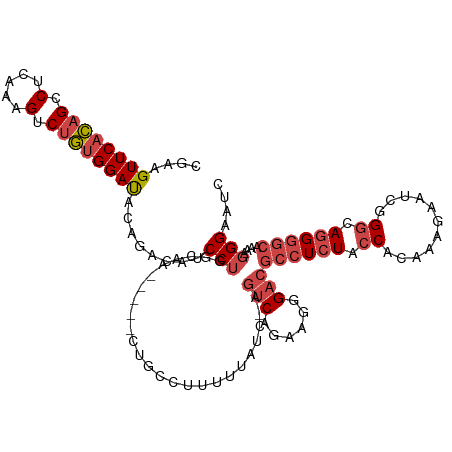
>dm3.chr2R 9555553 110 + 21146708 -AUCGAUAAGACAGGUUCCACAAAAGGUCUGCAUUGUGGGCGAAGUUCACAGACUCAAAGUCUGUGGAUACAGACACCCUGGCAG-----CUGUUAUUUUAUCAGUCAGAUGGGAC- -...((((((((((...((((((...(....).))))))((...((((((((((.....)))))))))).(((.....)))))..-----)))))...))))).(((......)))- ( -33.20, z-score = -1.60, R) >droAna3.scaffold_13266 16864141 117 + 19884421 AGCAAGUCAGACAGGUCCAGCAGCGCGUAUGCCUUGUCGGCCAAGUUCGUAGACUGAGUGACAAAGGACCUGCUGGUCCCAAAAAAGUCUCAGUCUGCAUGUUUAGCCGAAAGGGUA .((.....(((((((.((.((((.((....)).)))).))))......((((((((((.......(((((....))))).........)))))))))).)))))..((....)))). ( -42.39, z-score = -2.71, R) >droEre2.scaffold_4845 6335835 110 - 22589142 -AUCGAUAAGACAGGUUCCACAGAAGGUGCGCAUUGUGGGCGAAGUUCACAGCCUCAAAGUCUGUGGAUACGAACAGCCUGGCAA-----CUGCCUAUUUAUCAGUCAGAAAGGAC- -...((((((..((((((((((((...((.((..(((((((...)))))))))..))...))))))))........))))(((..-----..)))..)))))).(((......)))- ( -32.50, z-score = -1.26, R) >droYak2.chr2R 13746945 110 - 21139217 -AUCGAUAAGACAGGUUCCGCAGAAGGUCCGCAUUGUGGGUGAAGUUCACAGCCUCAAAGUCUGUGGACACAGACAGCCUGGUAA-----CUGUCUUUUCAUCAGUCAGAAAGGAC- -...((.(((((((..((((((((((((((((...))))(((.....))).)))).....))))))))..(((.....)))....-----))))))).))....(((......)))- ( -34.60, z-score = -1.52, R) >droSec1.super_1 7082352 110 + 14215200 -AUCGAUAAGACGGGUUCCACAGAAGGUCUGCAUUGUGGGCGAAGUUCACAGCCUCAAAGUCUGUGGAUGCAAACAUCCUGGCAG-----CUGCAUUUUUAUCAGUCAGAUGGGAC- -...(((((((((((.((((((((((((......(((((((...))))))))))).....)))))))).........))))((..-----..))..))))))).(((......)))- ( -30.40, z-score = 0.04, R) >droSim1.chr2R 8001628 110 + 19596830 -AUCGAUAAGACAGGUUCCACAGAAGGUCUGCAUUGUGGGCGAAGUUCACAGCCUCAAAGUCUGUGGAUGCAGACAUCCUGGCAG-----CUGCAUUUUUAUCAGUCAGAUGGGAC- -...(((((((..((((.(.(((...(((((((((((((((...))))))).((.((.....)).))))))))))...)))).))-----))....))))))).(((......)))- ( -32.40, z-score = -0.41, R) >consensus _AUCGAUAAGACAGGUUCCACAGAAGGUCUGCAUUGUGGGCGAAGUUCACAGCCUCAAAGUCUGUGGAUACAGACAUCCUGGCAA_____CUGCCUUUUUAUCAGUCAGAAAGGAC_ ....((((((.((((.(((((((...(....).)))))))....((((((((.(.....).))))))))........))))(((.......)))...)))))).(((......))). (-20.47 = -19.73 + -0.74)

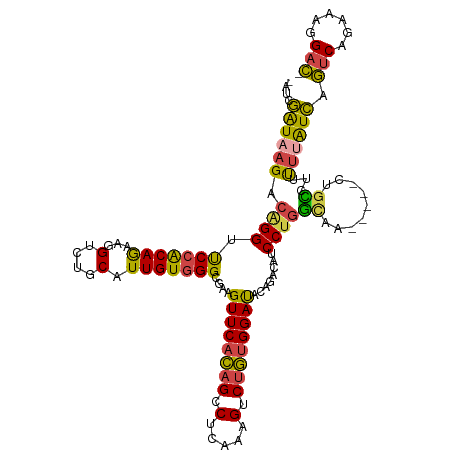
| Location | 9,555,592 – 9,555,703 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.64 |
| Shannon entropy | 0.43268 |
| G+C content | 0.50035 |
| Mean single sequence MFE | -34.51 |
| Consensus MFE | -18.58 |
| Energy contribution | -19.75 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9555592 111 + 21146708 CGAAGUUCACAGACUCAAAGUCUGUGGAUACAGACACCCUGGCAG-----CUGUUAUUUUAUC-AGUCAGAUGGGACGCCUCUACCAUAAAGAAUCGGGCAGGGGCAAAAGGGAAUC ....((((((((((.....)))))))))).......((((.....-----......(((((((-.....))))))).((((((.((...........)).))))))...)))).... ( -34.50, z-score = -2.25, R) >droAna3.scaffold_13266 16864181 117 + 19884421 CCAAGUUCGUAGACUGAGUGACAAAGGACCUGCUGGUCCCAAAAAAGUCUCAGUCUGCAUGUUUAGCCGAAAGGGUAAGCUGUACCACAAGGACAACGACAAGAAAACAAUGGAAAC (((..(((((((((((((.......(((((....))))).........))))))))))..(((((.((....)).)))))(((.((....))))).......))).....))).... ( -40.79, z-score = -4.31, R) >droEre2.scaffold_4845 6335874 111 - 22589142 CGAAGUUCACAGCCUCAAAGUCUGUGGAUACGAACAGCCUGGCAA-----CUGCCUAUUUAUC-AGUCAGAAAGGACGCCUCUACCACAAGGAAUCGGGCAGGGGCAGAAGGGAAUC ....((((((((.(.....).))))))))........(((.((..-----((((((.......-.(((......))).(((........)))....))))))..))...)))..... ( -32.30, z-score = -0.85, R) >droYak2.chr2R 13746984 111 - 21139217 UGAAGUUCACAGCCUCAAAGUCUGUGGACACAGACAGCCUGGUAA-----CUGUCUUUUCAUC-AGUCAGAAAGGACGCCUCUACCACAAGGAAUCGGGCAGGGGCAGAAGGGAAUC ...........(((((...((((((....)))))).(((((((..-----..((((((((...-.....)))))))).(((........))).))))))).)))))........... ( -39.30, z-score = -2.59, R) >droSec1.super_1 7082391 111 + 14215200 CGAAGUUCACAGCCUCAAAGUCUGUGGAUGCAAACAUCCUGGCAG-----CUGCAUUUUUAUC-AGUCAGAUGGGACGCCUCUACCACAAAGAAUCGGGCAGGGGCAAAAGGGAAUC ....((((((((.(.....).)))))))).......((((.((..-----..))..(((((((-.....))))))).((((((.((.(........))).))))))....))))... ( -29.00, z-score = 0.07, R) >droSim1.chr2R 8001667 111 + 19596830 CGAAGUUCACAGCCUCAAAGUCUGUGGAUGCAGACAUCCUGGCAG-----CUGCAUUUUUAUC-AGUCAGAUGGGACGCCUCUACCACAAAGAAUCGGGCAGGGGCAAAAGGGAAUC ....((((.(((((.....((((((....)))))).....))).(-----(((.((....)))-)))....))))))((((((.((.(........))).))))))........... ( -31.20, z-score = -0.29, R) >consensus CGAAGUUCACAGCCUCAAAGUCUGUGGAUACAGACAUCCUGGCAA_____CUGCCUUUUUAUC_AGUCAGAAGGGACGCCUCUACCACAAAGAAUCGGGCAGGGGCAAAAGGGAAUC ....((((((((.(.....).))))))))........(((.........................(((......)))((((((.((...........)).))))))....))).... (-18.58 = -19.75 + 1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:41 2011