| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,544,788 – 9,544,880 |
| Length | 92 |
| Max. P | 0.880920 |

| Location | 9,544,788 – 9,544,880 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 56.97 |
| Shannon entropy | 0.80808 |
| G+C content | 0.61235 |
| Mean single sequence MFE | -17.65 |
| Consensus MFE | -7.54 |
| Energy contribution | -7.98 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.880920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
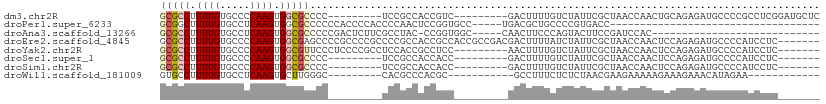
>dm3.chr2R 9544788 92 + 21146708 GCGCCUUUUGUGCCCCAAGUGGCGCCCC---------UCCGCCACCGUC---------GACUUUUGUCUAUUCGCUAACCAACUGCAGAGAUGCCCCGCCUCGGAUGCUC ((((.....)))).((..((((((....---------..))))))((((---------(((....))).....((.........))...)))).........))...... ( -21.80, z-score = -0.39, R) >droPer1.super_6233 176 70 + 1709 GCGGCUUUUGUGCCUCAAGUGGCGCCCCCCACCCCACCCCAACUCCGGUGCC-----UGACGCUGCCCCGUGACC----------------------------------- (((((....(((((......))))).........((((........))))..-----....))))).........----------------------------------- ( -19.60, z-score = -0.88, R) >droAna3.scaffold_13266 16850624 76 + 19884421 GCGCCUUUUGUGCCUCAAGUGGCGCCCCCGACUCUUCGCCUAC-CCGGUGGC-----CAACUUCCCAGUACUUCCGAUCCAC---------------------------- (((((.((((.....)))).)))))..........(((((...-..))))).-----.........................---------------------------- ( -16.40, z-score = -0.30, R) >droEre2.scaffold_4845 6325226 103 - 22589142 GCGCCUUUUGUGCCCCAAGUGGCGAGCCCCGCCCCGCCCCGCCACCGCCACCGCCGACGACUUUUAUCUAUUCGCUAACCAACUCCAGAGAUGCCCCAUCCUC------- ((((.....)))).....((((((.((........))..)))))).........................................((.((((...)))))).------- ( -16.10, z-score = 0.40, R) >droYak2.chr2R 13735963 94 - 21139217 GCGCCUUUUGUGCCCCAAGUGGCGUUCCCUCCCCGCCUCCACCGCCUCC---------AACUUUUGUCUAUUCGCUAACCAACUCCAGAGAUGCCCCAUCCUC------- (((((.((((.....)))).)))))........................---------............................((.((((...)))))).------- ( -10.50, z-score = 0.42, R) >droSec1.super_1 7071643 85 + 14215200 GCGCCUUUUGUGCCCCAAGUGGCGCCCC---------UCCGCCACCACC---------GACUUUUGUCUAUUCGCUAACCAACUCCAGAGAUGCCCCAUCCUC------- ((((.....)))).....((((((....---------..))))))....---------(((....)))..................((.((((...)))))).------- ( -16.90, z-score = -1.87, R) >droSim1.chr2R 7990911 85 + 19596830 GCGCCUUUUGUGCCCCAAGUGGCGCCCC---------UCCGCCACCACC---------GACUUUUGUCUAUUCGCUAACCAACUCCAGAGAUGCCCCAUCCUC------- ((((.....)))).....((((((....---------..))))))....---------(((....)))..................((.((((...)))))).------- ( -16.90, z-score = -1.87, R) >droWil1.scaffold_181009 2353762 78 - 3585778 GUGCCUUUUGUGCCUCAAGUGCUUGGGC---------CACGCCCACGC-----------GCCUUUCUCUCUAACGAAGAAAAAGAAAGAAACAUAGAA------------ ......((((((..((..((((.(((((---------...))))).))-----------))((((.((((....).))).))))...))..)))))).------------ ( -23.00, z-score = -2.40, R) >consensus GCGCCUUUUGUGCCCCAAGUGGCGCCCC_________UCCGCCACCGCC_________GACUUUUGUCUAUUCGCUAACCAACUCCAGAGAUGCCCCAUCCUC_______ (((((.((((.....)))).)))))..................................................................................... ( -7.54 = -7.98 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:40 2011