| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,543,198 – 9,543,296 |
| Length | 98 |
| Max. P | 0.984056 |

| Location | 9,543,198 – 9,543,296 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.17 |
| Shannon entropy | 0.34561 |
| G+C content | 0.46264 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -20.96 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.984056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
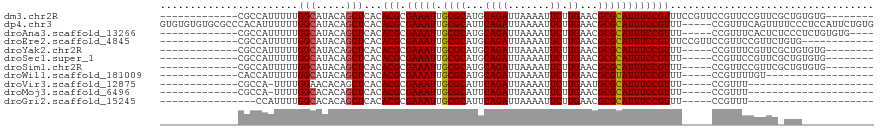
>dm3.chr2R 9543198 98 + 21146708 --------CACACAGCGAACGGAACGGAACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAAUGGCG------------- --------(((((.(((((..........((....))....(((((((...(((.......)))..)))))))..))))))))))........((((....)))).------------- ( -33.20, z-score = -2.36, R) >dp4.chr3 5237640 114 - 19779522 CACAGAAUGGAGGGAAAACUGAAACGG-----AAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGAAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAAAUGUGGGCGCACACACAC ...........(.((((.......((.-----...))....((((((((((.((.......)))))))))))).)))).)(((((.((..(((........)))..).).))))).... ( -34.30, z-score = -2.61, R) >droAna3.scaffold_13266 16849081 97 + 19884421 ----CACACAGAGGGAGAGUGAAACGG-----AAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAAUGGCG------------- ----...((((.......((((((((.-----...))....(((((((...(((.......)))..))))))).)))))).......))))..((((....)))).------------- ( -30.24, z-score = -1.98, R) >droEre2.scaffold_4845 6323654 94 - 22589142 ------------CACAGAACGGAACGGAACGGAAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAAUGGCG------------- ------------.((((..((..(((...((....))(((((((((((...(((.......)))..)))))).))))).))).))..))))..((((....)))).------------- ( -30.20, z-score = -2.04, R) >droYak2.chr2R 13734347 93 - 21139217 --------CACACAGCGAACGAAACGG-----AAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAAUGGCG------------- --------(((((.(((((.....((.-----...))....(((((((...(((.......)))..)))))))..))))))))))........((((....)))).------------- ( -33.20, z-score = -2.90, R) >droSec1.super_1 7070060 93 + 14215200 --------CACACAGCGAACGGAACGG-----AAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAAUGGCG------------- --------(((((.(((((.....((.-----...))....(((((((...(((.......)))..)))))))..))))))))))........((((....)))).------------- ( -33.20, z-score = -2.73, R) >droSim1.chr2R 7989324 93 + 19596830 --------CACACAGCGAACGGAACGG-----AAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAAUGGCG------------- --------(((((.(((((.....((.-----...))....(((((((...(((.......)))..)))))))..))))))))))........((((....)))).------------- ( -33.20, z-score = -2.73, R) >droWil1.scaffold_181009 2351701 83 - 3585778 ------------------ACAAAACGG-----AAACGGAAAUACGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAAUGGUG------------- ------------------......((.-----...))...(((((.((((.(((.......)))(((((((......))))))))))))))))((((....)))).------------- ( -22.50, z-score = -1.46, R) >droVir3.scaffold_12875 8143904 79 + 20611582 ---------------------AAACGG-----AAACGGAAAUGCGCAUUCAAGAAUUUUAAUCUGAAUGCGCAAUUUCGCGUGUGAGCUGUGUUCCAAAA-UGGCG------------- ---------------------...((.-----...))((((((((((((((.((.......))))))))))).)))))........(((((........)-)))).------------- ( -26.50, z-score = -2.92, R) >droMoj3.scaffold_6496 7762634 79 - 26866924 ---------------------AAACGG-----AAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGAAUGCGCAAUUUCGCGUGUGAGCUGUGUGCCAAAA-UGGCG------------- ---------------------..((((-----..(((((((((((((((((.((.......))))))))))).))))).))).....))))..(((....-.))).------------- ( -28.80, z-score = -3.10, R) >droGri2.scaffold_15245 1623436 77 + 18325388 ---------------------AAACGG-----AAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGAAUGCGCAAUUUCGCGUGUGAGCUGUGUGCCAAAAUGG---------------- ---------------------...((.-----...))((((((((((((((.((.......))))))))))).)))))(((..(.....)..)))........---------------- ( -24.90, z-score = -2.48, R) >consensus ____________CAG_GAACGAAACGG_____AAACGGAAAUGCGCGUUCAAGAAUUUUAAUCUGCAUGCGCAAUUUCGCGUGUGAGCUGUAUGCCAAAAAUGGCG_____________ .......................((((.......((((((((((((((...(((.......)))..)))))).))))).))).....))))..(((......))).............. (-20.96 = -21.22 + 0.26)

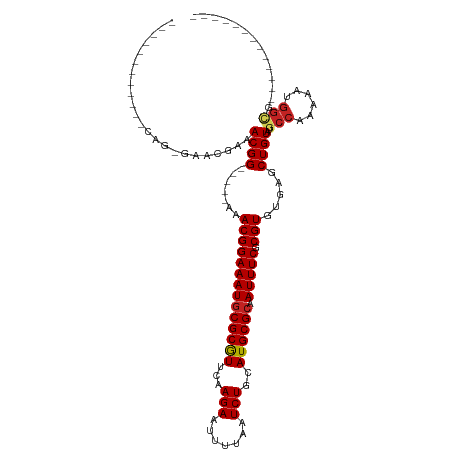

| Location | 9,543,198 – 9,543,296 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Shannon entropy | 0.34561 |
| G+C content | 0.46264 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -14.06 |
| Energy contribution | -14.07 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.689242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9543198 98 - 21146708 -------------CGCCAUUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUGCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUUCCGUUCCGUUCCGUUCGCUGUGUG-------- -------------.((((....))))(((((((....(((.(((((.((((...((((.......)).))...))))))))))))...((........))...))))))).-------- ( -28.50, z-score = -1.94, R) >dp4.chr3 5237640 114 + 19779522 GUGUGUGUGCGCCCACAUUUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUUCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUU-----CCGUUUCAGUUUUCCCUCCAUUCUGUG (((((((.(((((..........))).....)).)))))))(((((.((((.((((((.......)).)))).))))))))).....-----........................... ( -29.00, z-score = -2.40, R) >droAna3.scaffold_13266 16849081 97 - 19884421 -------------CGCCAUUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUGCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUU-----CCGUUUCACUCUCCCUCUGUGUG---- -------------.((((....))))((((((.....(((.(((((.((((...((((.......)).))...))))))))))))..-----..(.....).......)))))).---- ( -23.20, z-score = -1.73, R) >droEre2.scaffold_4845 6323654 94 + 22589142 -------------CGCCAUUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUGCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUUCCGUUCCGUUCCGUUCUGUG------------ -------------.((((....)))).(((((..(..(((.(((((.((((...((((.......)).))...))))))))))))...((...))....)..)))))------------ ( -22.10, z-score = -0.74, R) >droYak2.chr2R 13734347 93 + 21139217 -------------CGCCAUUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUGCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUU-----CCGUUUCGUUCGCUGUGUG-------- -------------.((((....))))(((((((..(((((.((((((((((...((((.......)).))...))))).....))))-----))))...))..))))))).-------- ( -28.50, z-score = -2.09, R) >droSec1.super_1 7070060 93 - 14215200 -------------CGCCAUUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUGCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUU-----CCGUUCCGUUCGCUGUGUG-------- -------------.((((....))))(((((((..(((((.((((((((((...((((.......)).))...))))).....))))-----))))...))..))))))).-------- ( -28.50, z-score = -2.16, R) >droSim1.chr2R 7989324 93 - 19596830 -------------CGCCAUUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUGCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUU-----CCGUUCCGUUCGCUGUGUG-------- -------------.((((....))))(((((((..(((((.((((((((((...((((.......)).))...))))).....))))-----))))...))..))))))).-------- ( -28.50, z-score = -2.16, R) >droWil1.scaffold_181009 2351701 83 + 3585778 -------------CACCAUUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUGCAGAUUAAAAUUCUUGAACGCGUAUUUCCGUUU-----CCGUUUUGU------------------ -------------..(((....))).(((.(((....(((.(((((.((((...((((.......)).))...))))))))))))..-----..))).)))------------------ ( -14.40, z-score = 0.42, R) >droVir3.scaffold_12875 8143904 79 - 20611582 -------------CGCCA-UUUUGGAACACAGCUCACACGCGAAAUUGCGCAUUCAGAUUAAAAUUCUUGAAUGCGCAUUUCCGUUU-----CCGUUU--------------------- -------------.....-....((((.((.((......))(((((.(((((((((((.......)).)))))))))))))).))))-----))....--------------------- ( -24.70, z-score = -3.70, R) >droMoj3.scaffold_6496 7762634 79 + 26866924 -------------CGCCA-UUUUGGCACACAGCUCACACGCGAAAUUGCGCAUUCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUU-----CCGUUU--------------------- -------------.(((.-....)))....(((....(((.(((((.((((.((((((.......)).)))).))))))))))))..-----..))).--------------------- ( -22.00, z-score = -2.71, R) >droGri2.scaffold_15245 1623436 77 - 18325388 ----------------CCAUUUUGGCACACAGCUCACACGCGAAAUUGCGCAUUCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUU-----CCGUUU--------------------- ----------------.......(((.....)))...(((.(((((.((((.((((((.......)).)))).))))))))))))..-----......--------------------- ( -18.20, z-score = -2.04, R) >consensus _____________CGCCAUUUUUGGCAUACAGCUCACACGCGAAAUUGCGCAUGCAGAUUAAAAUUCUUGAACGCGCAUUUCCGUUU_____CCGUUUCGUUC_CUG____________ .......................(((.....)))...(((.(((((.((((...((((.......)).))...)))))))))))).................................. (-14.06 = -14.07 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:39 2011