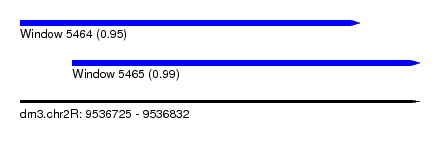
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,536,725 – 9,536,832 |
| Length | 107 |
| Max. P | 0.987845 |

| Location | 9,536,725 – 9,536,816 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.44 |
| Shannon entropy | 0.51372 |
| G+C content | 0.37339 |
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.39 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
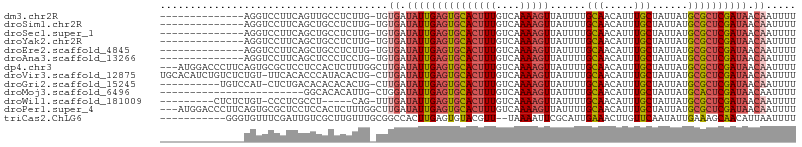
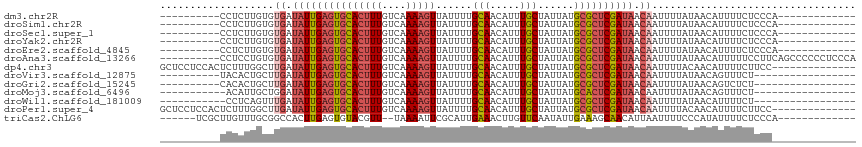
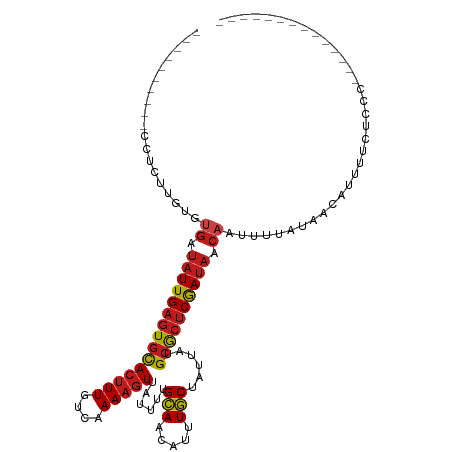
>dm3.chr2R 9536725 91 + 21146708 --------------AGGUCCUUCAGUUGCCUCUUG-UGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU --------------((((.........))))....-..((.(((((((((((((((....)))))......(((.....)))......)))))))))).))..... ( -19.80, z-score = -1.08, R) >droSim1.chr2R 7982873 91 + 19596830 --------------AGGUCCUUCAGCUGCCUCUUG-UGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU --------------((((.........))))....-..((.(((((((((((((((....)))))......(((.....)))......)))))))))).))..... ( -19.80, z-score = -0.83, R) >droSec1.super_1 7063574 91 + 14215200 --------------AGGUCCUUCAGCUGCCUCUUG-UGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU --------------((((.........))))....-..((.(((((((((((((((....)))))......(((.....)))......)))))))))).))..... ( -19.80, z-score = -0.83, R) >droYak2.chr2R 13727710 91 - 21139217 --------------AGGUCCUUCAGCUGCCUCUUG-UGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU --------------((((.........))))....-..((.(((((((((((((((....)))))......(((.....)))......)))))))))).))..... ( -19.80, z-score = -0.83, R) >droEre2.scaffold_4845 6313119 91 - 22589142 --------------AGGUCCUUCAGCUGCCUCUUG-UGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU --------------((((.........))))....-..((.(((((((((((((((....)))))......(((.....)))......)))))))))).))..... ( -19.80, z-score = -0.83, R) >droAna3.scaffold_13266 16842822 91 + 19884421 --------------AGGUCCUUCAGCUCCCUCCUG-UGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU --------------(((..............))).-..((.(((((((((((((((....)))))......(((.....)))......)))))))))).))..... ( -18.94, z-score = -1.28, R) >dp4.chr3 5231100 103 - 19779522 ---AUGGACCCUUCAGUGCGCUCCUCCACUCUUUGGCUUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU ---......((...((((........))))....)).(((.(((((((((((((((....)))))......(((.....)))......)))))))))).))).... ( -22.10, z-score = -1.15, R) >droVir3.scaffold_12875 8134003 104 + 20611582 UGCACAUCUGUCUCUGU-UUCACACCCAUACACUG-CUUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU .(((..........((.-....)).........))-)(((.(((((((((((((((....)))))......(((.....)))......)))))))))).))).... ( -19.31, z-score = -0.74, R) >droGri2.scaffold_15245 1615060 94 + 18325388 ----------UGUCCAU-CUCUGACACACACACUG-CUUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU ----------((((...-....)))).........-.(((.(((((((((((((((....)))))......(((.....)))......)))))))))).))).... ( -20.90, z-score = -1.92, R) >droMoj3.scaffold_6496 7751470 81 - 26866924 ------------------------GGCACACAUUG-CUGGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCACUCGAUAACAAUUUU ------------------------((((.....))-))...(((((((((((((((....)))))......(((.....)))......))))))))))........ ( -20.60, z-score = -2.51, R) >droWil1.scaffold_181009 1248593 90 + 3585778 ---------CUCUCUGU-CCCUCGCCU-----CAG-UUUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU ---------........-.........-----...-.(((.(((((((((((((((....)))))......(((.....)))......)))))))))).))).... ( -19.40, z-score = -2.40, R) >droPer1.super_4 3056832 103 + 7162766 ---AUGGACCCUUCAGUGCGCUCCUCCACUCUUUGGCUUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU ---......((...((((........))))....)).(((.(((((((((((((((....)))))......(((.....)))......)))))))))).))).... ( -22.10, z-score = -1.15, R) >triCas2.ChLG6 8385838 93 + 13544221 -----------GGGUGUUUCGAUUGUCGCUUGUUUGCGGCCACUUGAGUGUACGUU--UAAAAUUCGCAUUGAAACUUGUUCAAUAUUGAAAGCAACAUUAAUUUU -----------.(((((((((((....((..(((((((..(((....)))..))).--..))))..))((((((.....)))))))))))....))))))...... ( -16.40, z-score = 0.81, R) >consensus ______________AGGUCCUUCAGCUGCCUCUUG_UGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUU ......................................((.(((((((((((((((....)))))......(((.....)))......)))))))))).))..... (-16.04 = -16.39 + 0.36)
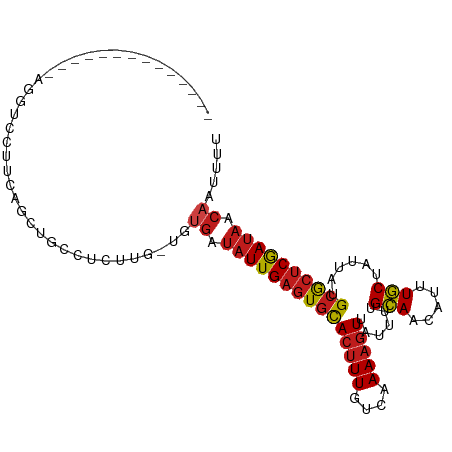
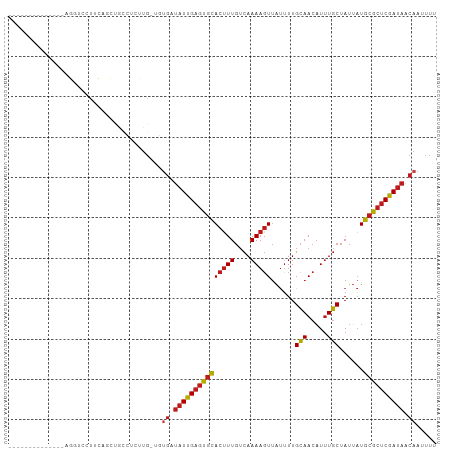
| Location | 9,536,739 – 9,536,832 |
|---|---|
| Length | 93 |
| Sequences | 13 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.05 |
| Shannon entropy | 0.36942 |
| G+C content | 0.33863 |
| Mean single sequence MFE | -18.56 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.39 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.987845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9536739 93 + 21146708 ----------CCUCUUGUGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUAUAACAUUUUCUCCCA------------- ----------.........((.(((((((((((((((....)))))......(((.....)))......)))))))))).)).....................------------- ( -18.70, z-score = -2.36, R) >droSim1.chr2R 7982887 93 + 19596830 ----------CCUCUUGUGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUAUAACAUUUUCUCCCA------------- ----------.........((.(((((((((((((((....)))))......(((.....)))......)))))))))).)).....................------------- ( -18.70, z-score = -2.36, R) >droSec1.super_1 7063588 93 + 14215200 ----------CCUCUUGUGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUAUAACAUUUUCUCCCA------------- ----------.........((.(((((((((((((((....)))))......(((.....)))......)))))))))).)).....................------------- ( -18.70, z-score = -2.36, R) >droYak2.chr2R 13727724 93 - 21139217 ----------CCUCUUGUGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUAUAACAUUUUCUCCCA------------- ----------.........((.(((((((((((((((....)))))......(((.....)))......)))))))))).)).....................------------- ( -18.70, z-score = -2.36, R) >droEre2.scaffold_4845 6313133 93 - 22589142 ----------CCUCUUGUGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUAUAACAUUUUCUCCCA------------- ----------.........((.(((((((((((((((....)))))......(((.....)))......)))))))))).)).....................------------- ( -18.70, z-score = -2.36, R) >droAna3.scaffold_13266 16842836 106 + 19884421 ----------CCUCCUGUGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUAUAACAUUUUUCCUUCAGCCCCCCUCCCA ----------.........((.(((((((((((((((....)))))......(((.....)))......)))))))))).)).................................. ( -18.70, z-score = -2.59, R) >dp4.chr3 5231116 102 - 19779522 GCUCCUCCACUCUUUGGCUUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUACAACAUUUUCUUCC-------------- ......(((.....))).(((.(((((((((((((((....)))))......(((.....)))......)))))))))).)))...................-------------- ( -19.80, z-score = -2.34, R) >droVir3.scaffold_12875 8134030 89 + 20611582 ----------UACACUGCUUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUAUAACAGUUUCU----------------- ----------........(((.(((((((((((((((....)))))......(((.....)))......)))))))))).)))................----------------- ( -19.30, z-score = -1.85, R) >droGri2.scaffold_15245 1615077 89 + 18325388 ----------CACACUGCUUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUAUAACAGUCUCU----------------- ----------........(((.(((((((((((((((....)))))......(((.....)))......)))))))))).)))................----------------- ( -19.30, z-score = -1.77, R) >droMoj3.scaffold_6496 7751475 88 - 26866924 -----------ACAUUGCUGGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCACUCGAUAACAAUUUUAUAACAGUUUCU----------------- -----------.....((((..(((((((((((((((....)))))......(((.....)))......))))))))))............))))....----------------- ( -18.64, z-score = -1.76, R) >droWil1.scaffold_181009 1248607 88 + 3585778 -----------CCUCAGUUUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUAUAACAUUUUCU----------------- -----------.......(((.(((((((((((((((....)))))......(((.....)))......)))))))))).)))................----------------- ( -19.40, z-score = -2.71, R) >droPer1.super_4 3056848 102 + 7162766 GCUCCUCCACUCUUUGGCUUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUACAACAUUUUCUUCC-------------- ......(((.....))).(((.(((((((((((((((....)))))......(((.....)))......)))))))))).)))...................-------------- ( -19.80, z-score = -2.34, R) >triCas2.ChLG6 8385852 95 + 13544221 ------UCGCUUGUUUGCGGCCACUUGAGUGUACGUU--UAAAAUUCGCAUUGAAACUUGUUCAAUAUUGAAAGCAACAUUAAUUUUCCCAUAUUUUCUCCCA------------- ------..((((....(((..(((....)))..))).--......(((.((((((.....))))))..)))))))............................------------- ( -12.90, z-score = 0.19, R) >consensus __________CCUCUUGUGUGAUAUUGAGUGCACUUUGUCAAAAGUUAUUUUGCAACAUUUGCUAUUAUGCGCUCGAUAACAAUUUUAUAACAUUUUCUCCC______________ ...................((.(((((((((((((((....)))))......(((.....)))......)))))))))).)).................................. (-16.04 = -16.39 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:37 2011