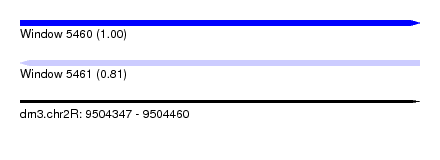
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,504,347 – 9,504,460 |
| Length | 113 |
| Max. P | 0.995798 |

| Location | 9,504,347 – 9,504,460 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.64 |
| Shannon entropy | 0.41541 |
| G+C content | 0.37431 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -15.88 |
| Energy contribution | -18.04 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.995798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9504347 113 + 21146708 -AAGUUCGAAUACGUAAUUAUUCGUAAAACACAAAGGCUUGAUAACAGUCAAUGUAUAUGUAUAUGUAUUGUAUGCAUACACCGAGCAGAGCGGAGCCUUUGAUAACUUAACCA -(((((((((((......)))))).......((((((((((((....)))).....(((((((((.....)))))))))..(((.......)))))))))))..)))))..... ( -27.70, z-score = -2.29, R) >droSim1.chr2R 7948917 108 + 19596830 GAAAAGCGAAUACGUAAUUAUUCGUAAAACACAAAGGCUUGAUA----UAUAUA-GUAUGUAUAUAUAUAGUAUGUAUACAC-GAGCAGCGCAGCGCCUUUGAUAACUUAACCA .....(((((((......)))))))......(((((((.((.((----((((((-.(((((....))))).)))))))))).-..((......)))))))))............ ( -27.80, z-score = -3.10, R) >droSec1.super_1 7021982 108 + 14215200 GAAAAGCGAAUACGUAAAUAUUCGUAAAACACAAAGGCUUGAUA----UAUAUA-GUAUGUAUAUAUAUAGUAUGUAUACAC-GAGCAGCGCAGAGCCUUUGAUAACUUAACCA .....(((((((......)))))))......(((((((((..((----((((((-.(((((....))))).))))))))...-..((...)).)))))))))............ ( -27.90, z-score = -3.81, R) >droYak2.chr2R 13695242 97 - 21139217 GAGAAGCGAAUACGUAAUUAUUCGCAAAACACAAAGGCUUCGGAG----------AAGUGAGUAUAUA-------CACGAGCAGAGCAGAGCAGAGCCUUUGAUAACUUAACCA (((..(((((((......)))))))......(((((((((.....----------..(((........-------)))..((........)).)))))))))....)))..... ( -24.70, z-score = -3.54, R) >droEre2.scaffold_4845 6278461 96 - 22589142 -GAAAGCGAAUACGUAAUUAUUCGUAAAACACAAAGGCUGGCAAG----------AAGUGCAUAUUUA-------CACGGGCAGCGAAGAGCCGCGCCUUUGAUAACUUAACCA -....(((((((......)))))))......(((((((..((...----------..(((........-------)))..)).(((......))))))))))............ ( -24.30, z-score = -1.67, R) >consensus GAAAAGCGAAUACGUAAUUAUUCGUAAAACACAAAGGCUUGAUA____U__AU__AUAUGUAUAUAUAU_GUAUGCAUACAC_GAGCAGAGCAGAGCCUUUGAUAACUUAACCA .....(((((((......)))))))......(((((((((.................((((((((.....)))))))).......((...)).)))))))))............ (-15.88 = -18.04 + 2.16)

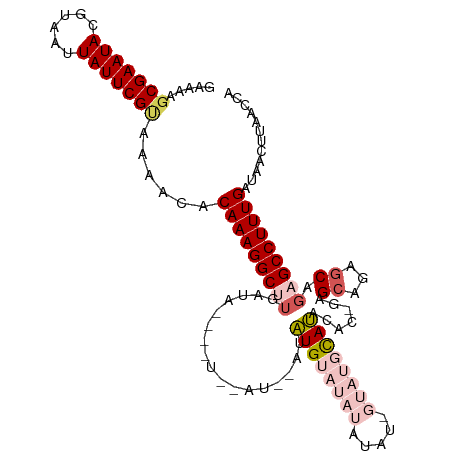

| Location | 9,504,347 – 9,504,460 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.64 |
| Shannon entropy | 0.41541 |
| G+C content | 0.37431 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -14.62 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.805931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
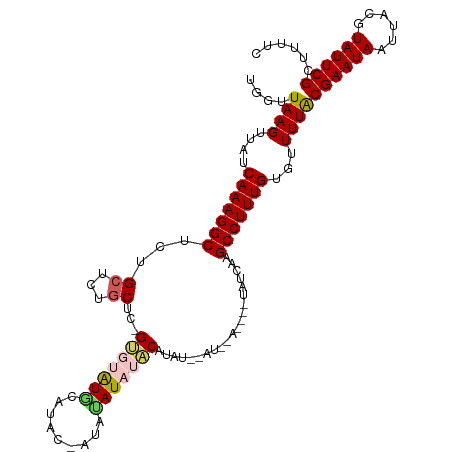
>dm3.chr2R 9504347 113 - 21146708 UGGUUAAGUUAUCAAAGGCUCCGCUCUGCUCGGUGUAUGCAUACAAUACAUAUACAUAUACAUUGACUGUUAUCAAGCCUUUGUGUUUUACGAAUAAUUACGUAUUCGAACUU- .....(((((..((((((((.........((((((((((.................)))))))))).........)))))))).......((((((......)))))))))))- ( -23.80, z-score = -1.07, R) >droSim1.chr2R 7948917 108 - 19596830 UGGUUAAGUUAUCAAAGGCGCUGCGCUGCUC-GUGUAUACAUACUAUAUAUAUACAUAC-UAUAUA----UAUCAAGCCUUUGUGUUUUACGAAUAAUUACGUAUUCGCUUUUC ....((((....(((((((..(((((.....-)))))........(((((((((.....-))))))----)))...)))))))...))))((((((......))))))...... ( -22.30, z-score = -0.85, R) >droSec1.super_1 7021982 108 - 14215200 UGGUUAAGUUAUCAAAGGCUCUGCGCUGCUC-GUGUAUACAUACUAUAUAUAUACAUAC-UAUAUA----UAUCAAGCCUUUGUGUUUUACGAAUAUUUACGUAUUCGCUUUUC ....((((....((((((((.(((((.....-)))))........(((((((((.....-))))))----)))..))))))))...))))(((((((....)))))))...... ( -23.40, z-score = -1.89, R) >droYak2.chr2R 13695242 97 + 21139217 UGGUUAAGUUAUCAAAGGCUCUGCUCUGCUCUGCUCGUG-------UAUAUACUCACUU----------CUCCGAAGCCUUUGUGUUUUGCGAAUAAUUACGUAUUCGCUUCUC ............(((((((((.((........))..(((-------........)))..----------....).))))))))......(((((((......)))))))..... ( -21.30, z-score = -2.02, R) >droEre2.scaffold_4845 6278461 96 + 22589142 UGGUUAAGUUAUCAAAGGCGCGGCUCUUCGCUGCCCGUG-------UAAAUAUGCACUU----------CUUGCCAGCCUUUGUGUUUUACGAAUAAUUACGUAUUCGCUUUC- ....((((....((((((((((((.....)))))..(((-------((....)))))..----------.......)))))))...))))((((((......)))))).....- ( -26.10, z-score = -1.99, R) >consensus UGGUUAAGUUAUCAAAGGCUCUGCUCUGCUC_GUGUAUGCAUAC_AUAUAUAUACAUAU__AU__A____UAUCAAGCCUUUGUGUUUUACGAAUAAUUACGUAUUCGCUUUUC ....((((....(((((((...((...))...(((((((.........))))))).....................)))))))...))))((((((......))))))...... (-14.62 = -15.10 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:33 2011