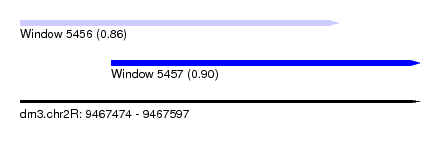
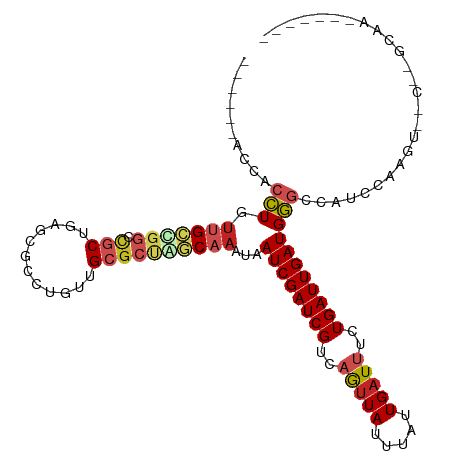
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,467,474 – 9,467,597 |
| Length | 123 |
| Max. P | 0.901573 |

| Location | 9,467,474 – 9,467,572 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.99 |
| Shannon entropy | 0.59282 |
| G+C content | 0.47350 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9467474 98 + 21146708 GACCUGACCACCUGUUGCUGGGCGCUGAGCGCCUGUUGCGCUAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGCCAACCAAGU---------------- ..............(((...((((((.(((((.....)))))))).....(((((((((((((.....))))))....)))))))..)))...)))..---------------- ( -27.10, z-score = -0.69, R) >dp4.chr3 5163505 101 - 19779522 ------GCCACCUGUUGUCGACUGCUGAGCGCCUGUUGCGCUGACAAAUAAUCGAUCGUAUGUUAUUUAUUGAUUUCUGAUUGAUGGGACCAUCCGCCAACUGUCAA------- ------....(((.(((((..(....)(((((.....))))))))))...((((((((...((((.....))))...)))))))))))...................------- ( -21.20, z-score = 0.14, R) >droAna3.scaffold_13266 16769769 87 + 19884421 -------CCACCUGUUGCCGGUCGCUGAGCGCCUGAUGCGCUGGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGCAGGCC-------------------- -------...(((((((((((.(((............)))))))).....(((((((((((((.....))))))....))))))).))))))..-------------------- ( -29.20, z-score = -1.45, R) >droEre2.scaffold_4845 6242247 95 - 22589142 UGUUUGACCACCUGUUGCCGGGCUCUGAGCGCCUGUUGCGCCAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGG--CAUCCAUU----------------- ((..((.((..(((.((((((((.......)))))..))).)))......(((((((((((((.....))))))....)))))))))--))..))..----------------- ( -25.20, z-score = -0.82, R) >droYak2.chr2R 13657948 101 - 21139217 ------ACCACCUGUUGCCGGGCUCUGAGCGCCUGUUGCGCUAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGCCAUC-AAGUCCCCAGCAAA------ ------......(((((..(((((.(((..((((.((((....))))...(((((((((((((.....))))))....)))))))))))..))-)))))).)))))..------ ( -31.50, z-score = -2.05, R) >droSec1.super_1 6985735 106 + 14215200 ----UGACCACCUGUUGCCGGGCGCUGAGCGCCUGUUGCGCUAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGCCAUCCAAGUUCCCCGCCAAAG---- ----..........(((.((((.(((.(((((.....)))))))).....(((((((((((((.....))))))....)))))))((.....))......)))).)))..---- ( -29.10, z-score = -0.72, R) >droSim1.chr2R 7911013 106 + 19596830 ----UGACCACCUGUUGCUGGGCGCUGAGCGCCUGUUGCGCUAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGCCAUCCAAGUCCCCCGCCAAAG---- ----..........(((.((((.(((.(((((.....)))))))).....(((((((((((((.....))))))....)))))))((((.......)))))))).)))..---- ( -30.00, z-score = -1.03, R) >droPer1.super_4 2990434 101 + 7162766 ------UCCACCUGUUGUCGACUGCUGAGCGCCUGUUGCGCUGACAAAUAAUCGAUCGUAUGUUAUUUAUUGAUUUCUGAUUGAUGGGACCAUCCGCCAACUGUCAA------- ------....(((.(((((..(....)(((((.....))))))))))...((((((((...((((.....))))...)))))))))))...................------- ( -21.20, z-score = -0.05, R) >droVir3.scaffold_12875 9480846 106 - 20611582 -------CAGCUUGUCGCCGGCUGCUGGGCGCCUGUUGCGCAGCCAAAUAAUCGAUCGCAAAUUAUUUAUUGAUUUUUGAUUGAUGAGCCACAGAGUAUU-AUGUAAUAUGUAU -------..((((((.(((((((((..(((....)))..)))))).....((((((((.((((((.....)))))).))))))))).)).))).)))...-............. ( -28.80, z-score = -0.59, R) >droMoj3.scaffold_6496 15797058 101 + 26866924 ------GUCGCUUGUAGCUGGCUGCUGGGCGCCUGUUGCCCACUCAAAUAAUCGAUCGCAAAUUAUUUAUUGAUUUUUGAUUGAUGAGCCUCAGGUGAUUUACGCAA------- ------((((((((.....((((..((((((.....))))))........((((((((.((((((.....)))))).)))))))).)))).))))))))........------- ( -33.10, z-score = -2.68, R) >droGri2.scaffold_15245 13605256 106 + 18325388 -----GUUUCGUUGCUGUUGGCUGCUGGGCGCCUGUUGCGCUGACAAAUAAUCGAUCGCAAAGUAUUUAUUGAUUUUUGAUUGAUGAGCAGCGGCGAGAACAAAAAUUGAC--- -----.(((((((((((((...((.(.(((((.....))))).)))....((((((((.(((((........))))))))))))).)))))))))))))............--- ( -34.00, z-score = -2.12, R) >consensus ______ACCACCUGUUGCCGGCCGCUGAGCGCCUGUUGCGCUAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGCCAUCCAAGU__C__GCAA_______ ..........(((.(((((((.(((............))))))))))...((((((((..(((((.....)))))..))))))))))).......................... (-15.28 = -15.85 + 0.57)

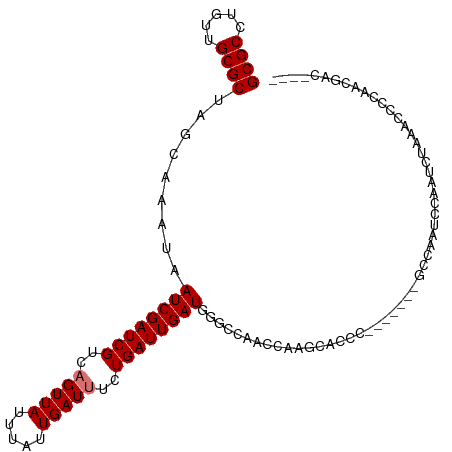
| Location | 9,467,502 – 9,467,597 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.57 |
| Shannon entropy | 0.62240 |
| G+C content | 0.46322 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -13.82 |
| Energy contribution | -14.07 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9467502 95 + 21146708 GCGCCUGUUGCGCUAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGCCAACCAAGU------------CCCAUCCAAUUUGAAUCCAAACGAC---- ((((.....))))..........(((...((((((.....))))))(((.((((((((((.(......).------------))))).))))).)))......))).---- ( -22.70, z-score = -1.78, R) >dp4.chr3 5163527 104 - 19779522 GCGCCUGUUGCGCUGACAAAUAAUCGAUCGUAUGUUAUUUAUUGAUUUCUGAUUGAUGGGACCAUCCGCCAACU-------GUCAAGCCAGUCAAUACAUUACGGGGCGAU ((.(((((((.(((((((....((((((((...((((.....))))...)))))))).((.....))......)-------))).)))))((....))...)))))))... ( -25.30, z-score = 0.02, R) >droAna3.scaffold_13266 16769790 89 + 19884421 GCGCCUGAUGCGCUGGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGC-----AGGCCC----------UCGAUGCAUGAGAGCCACCAAG------- ..((((..(((....)))....(((((((((((((.....))))))....)))))))))))-----.(((.(----------(((.....)))).)))......------- ( -26.30, z-score = -0.71, R) >droEre2.scaffold_4845 6242275 91 - 22589142 GCGCCUGUUGCGCCAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGCAUCCAUUGGCAUUCAUGCUUUGCCAAGCGAC-------------------- (((((..((((....))))...(((((((((((((.....))))))....)))))))))).....((((((.........))))))))...-------------------- ( -27.50, z-score = -1.70, R) >droYak2.chr2R 13657970 108 - 21139217 GCGCCUGUUGCGCUAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGCCAUC-AAGUCCCCAGC-AAAGGCAUUCCACUCGAUGCCCCAAAAACGGC- ..(((..((((....))))...(((((((((((((.....))))))....)))))))(((.((((-.(((.....((-....)).....))).)))).)))......)))- ( -28.90, z-score = -1.28, R) >droSec1.super_1 6985759 107 + 14215200 GCGCCUGUUGCGCUAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGCCAUCCAAGUUCCCCGCCAAAGCCCAUCCAAUUUGAAUCCCAACGAC---- ((((.....))))..........(((...((((((.....))))))(((.(((((((((((....................)))))).))))).)))......))).---- ( -24.15, z-score = -1.52, R) >droSim1.chr2R 7911037 107 + 19596830 GCGCCUGUUGCGCUAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGCCAUCCAAGUCCCCCGCCAAAGCCCAUCCAAUUUGAAUCCCAACGAC---- ((((.....))))..........(((...((((((.....))))))(((.(((((((((((....................)))))).))))).)))......))).---- ( -24.15, z-score = -1.61, R) >droPer1.super_4 2990456 104 + 7162766 GCGCCUGUUGCGCUGACAAAUAAUCGAUCGUAUGUUAUUUAUUGAUUUCUGAUUGAUGGGACCAUCCGCCAACU-------GUCAAGCCAGUCAAUACAUUACGGGGCGAU ((.(((((((.(((((((....((((((((...((((.....))))...)))))))).((.....))......)-------))).)))))((....))...)))))))... ( -25.30, z-score = 0.02, R) >consensus GCGCCUGUUGCGCUAGCAAAUAAUCGAUCGUCAGUUAUUUAUUGAUUUCUGAUUGAUGGGCCAACCAAGCACCC_______GCCAAUCCAAUCUAAACCCCAACGAC____ ((((.....)))).........((((((((..(((((.....)))))..))))))))...................................................... (-13.82 = -14.07 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:30 2011