| Sequence ID | dm3.chr2R |
|---|---|
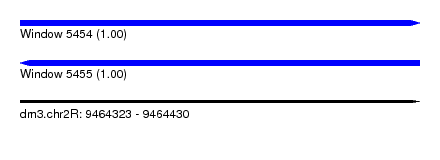
| Location | 9,464,323 – 9,464,430 |
| Length | 107 |
| Max. P | 0.999233 |

| Location | 9,464,323 – 9,464,430 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.66 |
| Shannon entropy | 0.32597 |
| G+C content | 0.41778 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -24.41 |
| Energy contribution | -24.21 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.91 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
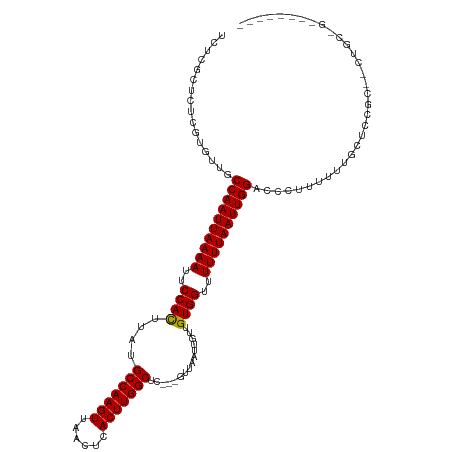
>dm3.chr2R 9464323 107 + 21146708 UCUCGCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUC---GUUAAUUGUUGUGGCUUUUUAAUUGGACCCUUUUUUUCUCCUC--CUGCUG-------- ...(((....)))...((((((((((..((((....(((((((......))))))).(---(.....))..))))..)))))))))).................--......-------- ( -24.90, z-score = -4.66, R) >droSim1.chr2R 7907697 107 + 19596830 UCUCGCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUC---GUUAAUUGUUGUGGCUUUUUAAUUGGACCCUUUUUUUCUCCUC--CUGCUG-------- ...(((....)))...((((((((((..((((....(((((((......))))))).(---(.....))..))))..)))))))))).................--......-------- ( -24.90, z-score = -4.66, R) >droSec1.super_1 6982407 106 + 14215200 UCUCGCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUC---GUUAAUUGUUGUGGCUUUUUAAUUGGACCCUUUUUU-CUCCUC--CUGAUG-------- ...(((....)))...((((((((((..((((....(((((((......))))))).(---(.....))..))))..))))))))))..........-......--......-------- ( -24.90, z-score = -4.38, R) >droEre2.scaffold_4845 6238821 110 - 22589142 UCUCGCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUC---GUUAAUUGUUGUGGCUUUUUAAUUGGACCCUUUUUUUCUCCUC--CUGCUGCUG----- ...(((....)))...((((((((((..((((....(((((((......))))))).(---(.....))..))))..)))))))))).................--.........----- ( -24.90, z-score = -4.12, R) >droAna3.scaffold_13266 16766370 107 + 19884421 CCUCUCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUC---GUUAAUUGUUGUGGCUUUUUAAUUGGACCCUUUUUUUCUCCUG--CGCCCA-------- ..........((((..((((((((((..((((....(((((((......))))))).(---(.....))..))))..))))))))))................)--)))...-------- ( -25.67, z-score = -4.59, R) >dp4.chr3 5160301 118 - 19779522 --UCUCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUCCCCGUUAAUUGUCGUGGUUUUUUAAUUGGACCCUUUUUUGCUGCGGGGCUGCCGGGCUGCCG --.....((((.((.(((((((((((..((((....(((((((......))))))).....(........)))))..)))))))))..(((...........))))).))))))...... ( -33.10, z-score = -1.85, R) >droPer1.super_4 2987145 118 + 7162766 --UCUCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUCCCCGUUAAUUGUCGUGGUUUUUUAAUUGGACCCUUUUUUGCUGCGGGGCUGCCGGGCUGCCG --.....((((.((.(((((((((((..((((....(((((((......))))))).....(........)))))..)))))))))..(((...........))))).))))))...... ( -33.10, z-score = -1.85, R) >droWil1.scaffold_180701 3811851 95 + 3904529 ----GCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUC---GUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUUUUGCUC------------------ ----((..........((((((((((..((((....(((((((......))))))).(---(.....))..))))..))))))))))...........))..------------------ ( -26.60, z-score = -4.90, R) >droMoj3.scaffold_6496 15793674 103 + 26866924 UGCCGCUCUUAUGUUGCCAAUUAAAAUUCCAUUUAUGCCAAGUUAACUCACUUGGCUC---GUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUCGGUGCUGCUG-------------- .((.((..........((((((((((..((((....(((((((......)))))))..---..........))))..))))))))))(((......))))).))..-------------- ( -29.59, z-score = -4.07, R) >droVir3.scaffold_12875 9477904 100 - 20611582 ---CGCUCUUAUGUUGCCAAUUAAAAUUCCAUUUAUGCCAAGUUAACUCACUUGGCUC---GUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUCGGAGCUGAUG-------------- ---.(((((.......((((((((((..((((....(((((((......)))))))..---..........))))..)))))))))).........))))).....-------------- ( -30.38, z-score = -4.77, R) >droGri2.scaffold_15245 13602157 106 + 18325388 UGUCGCUCUCAUGUUGCCAAUUAAAAUUCCAUUUAUGCCAAGUUAACUCACUUGGCUC---GUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUCGGUGCUGGUGGUG----------- ...((((..((.((..((((((((((..((((....(((((((......)))))))..---..........))))..))))))))))(((......)))))))..))))----------- ( -29.49, z-score = -3.14, R) >consensus UCUCGCUCUCGUGUUGCCAAUUAAAAUUCCACUUAUGCCAAGUUAACUCACUUGGCUC___GUUAAUUGUUGUGGUUUUUUAAUUGGACCCUUUUUUGCUCCGC__CUGC_G________ ................((((((((((..((((....(((((((......)))))))...............))))..))))))))))................................. (-24.41 = -24.21 + -0.20)

| Location | 9,464,323 – 9,464,430 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.66 |
| Shannon entropy | 0.32597 |
| G+C content | 0.41778 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.67 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.998425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
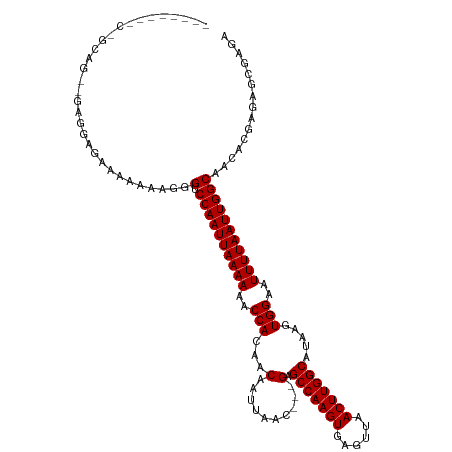
>dm3.chr2R 9464323 107 - 21146708 --------CAGCAG--GAGGAGAAAAAAAGGGUCCAAUUAAAAAGCCACAACAAUUAAC---GAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGCGAGA --------......--...............(.((((((((((..((((..(.......---).(((((((......)))))))....))))..)))))))))))..(.(....).)... ( -27.50, z-score = -3.88, R) >droSim1.chr2R 7907697 107 - 19596830 --------CAGCAG--GAGGAGAAAAAAAGGGUCCAAUUAAAAAGCCACAACAAUUAAC---GAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGCGAGA --------......--...............(.((((((((((..((((..(.......---).(((((((......)))))))....))))..)))))))))))..(.(....).)... ( -27.50, z-score = -3.88, R) >droSec1.super_1 6982407 106 - 14215200 --------CAUCAG--GAGGAG-AAAAAAGGGUCCAAUUAAAAAGCCACAACAAUUAAC---GAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGCGAGA --------......--......-........(.((((((((((..((((..(.......---).(((((((......)))))))....))))..)))))))))))..(.(....).)... ( -27.50, z-score = -3.77, R) >droEre2.scaffold_4845 6238821 110 + 22589142 -----CAGCAGCAG--GAGGAGAAAAAAAGGGUCCAAUUAAAAAGCCACAACAAUUAAC---GAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGCGAGA -----.........--...............(.((((((((((..((((..(.......---).(((((((......)))))))....))))..)))))))))))..(.(....).)... ( -27.50, z-score = -3.43, R) >droAna3.scaffold_13266 16766370 107 - 19884421 --------UGGGCG--CAGGAGAAAAAAAGGGUCCAAUUAAAAAGCCACAACAAUUAAC---GAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGAGAGG --------.((((.--(............).))))((((((((..((((..(.......---).(((((((......)))))))....))))..))))))))(....).(....)..... ( -27.40, z-score = -3.35, R) >dp4.chr3 5160301 118 + 19779522 CGGCAGCCCGGCAGCCCCGCAGCAAAAAAGGGUCCAAUUAAAAAACCACGACAAUUAACGGGGAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGAGA-- ((...((((.((.((...)).))......))))((((((((((..((((..(........)...(((((((......)))))))....))))..)))))))))).....)).......-- ( -32.30, z-score = -1.44, R) >droPer1.super_4 2987145 118 - 7162766 CGGCAGCCCGGCAGCCCCGCAGCAAAAAAGGGUCCAAUUAAAAAACCACGACAAUUAACGGGGAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGAGA-- ((...((((.((.((...)).))......))))((((((((((..((((..(........)...(((((((......)))))))....))))..)))))))))).....)).......-- ( -32.30, z-score = -1.44, R) >droWil1.scaffold_180701 3811851 95 - 3904529 ------------------GAGCAAAAAAAGGGUCCAAUUAAAAAACCACAACAAUUAAC---GAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGC---- ------------------.............(.((((((((((..((((..(.......---).(((((((......)))))))....))))..)))))))))))...........---- ( -25.50, z-score = -4.15, R) >droMoj3.scaffold_6496 15793674 103 - 26866924 --------------CAGCAGCACCGAAAAGGGUCCAAUUAAAAAACCACAACAAUUAAC---GAGCCAAGUGAGUUAACUUGGCAUAAAUGGAAUUUUAAUUGGCAACAUAAGAGCGGCA --------------..((.((.((.....))(.((((((((((..(((...(.......---).(((((((......))))))).....)))..))))))))))).........)).)). ( -29.00, z-score = -4.09, R) >droVir3.scaffold_12875 9477904 100 + 20611582 --------------CAUCAGCUCCGAAAAGGGUCCAAUUAAAAAACCACAACAAUUAAC---GAGCCAAGUGAGUUAACUUGGCAUAAAUGGAAUUUUAAUUGGCAACAUAAGAGCG--- --------------.....((((........(.((((((((((..(((...(.......---).(((((((......))))))).....)))..))))))))))).......)))).--- ( -27.46, z-score = -4.40, R) >droGri2.scaffold_15245 13602157 106 - 18325388 -----------CACCACCAGCACCGAAAAGGGUCCAAUUAAAAAACCACAACAAUUAAC---GAGCCAAGUGAGUUAACUUGGCAUAAAUGGAAUUUUAAUUGGCAACAUGAGAGCGACA -----------........((.((.....))(.((((((((((..(((...(.......---).(((((((......))))))).....)))..))))))))))).........)).... ( -26.00, z-score = -3.16, R) >consensus ________C_GCAG__GAGGAGAAAAAAAGGGUCCAAUUAAAAAACCACAACAAUUAAC___GAGCCAAGUGAGUUAACUUGGCAUAAGUGGAAUUUUAAUUGGCAACACGAGAGCGAGA ...............................(.((((((((((..(((......((......))(((((((......))))))).....)))..)))))))))))............... (-21.67 = -21.67 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:28 2011