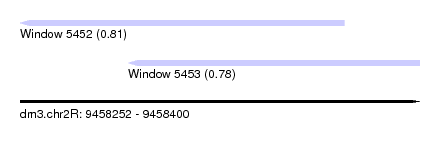
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,458,252 – 9,458,400 |
| Length | 148 |
| Max. P | 0.809446 |

| Location | 9,458,252 – 9,458,372 |
|---|---|
| Length | 120 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.12 |
| Shannon entropy | 0.54065 |
| G+C content | 0.44493 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -13.27 |
| Energy contribution | -12.86 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.809446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
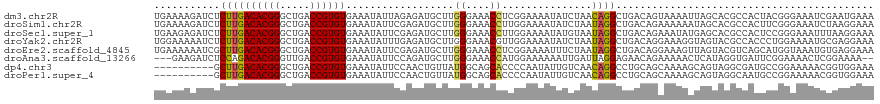
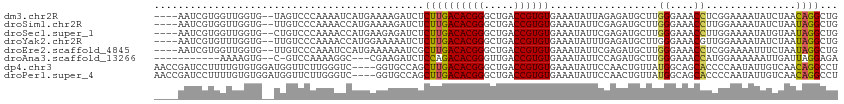
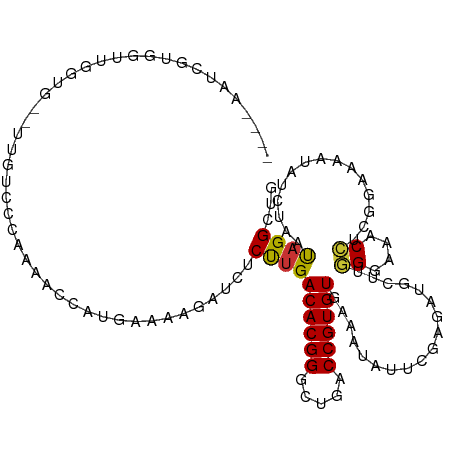
>dm3.chr2R 9458252 120 - 21146708 UGAAAAGAUCUCUUGACACGGGCUGACCGUGUGAAAUAUUAGAGAUGCUUGGGAAACCUCGGAAAAUAUCUAACAGGCUGACAGUAAAAUUAGCACGCCACUACGGGAAAUCGAAUGAAA ......(((.(((((((((((.....))))))......(((((.((.((.((....))..))...)).)))))..(((((..(((...)))..)).)))....))))).)))........ ( -28.30, z-score = -1.73, R) >droSim1.chr2R 7901759 120 - 19596830 UGAAAAGAUCUCUUGACACGGGCUGACCGUGUGAAAUAUUCGAGAUGCUUGGGAAACCUUGGAAAAUAUCUAAUAGGCUGACAGAAAAAAUAGCACGCCACUUCGGGAAAUCUAAGGAAA ....((((((((((.((((((.....)))))).))......))))).)))......(((((((............(((((.(..........))).))).(....)....)))))))... ( -29.50, z-score = -1.88, R) >droSec1.super_1 6976355 120 - 14215200 UGAAGAGAUCUCUUGACACGGGCUGACCGUGUGAAAUAUUCGAGAUGCUUGGGAAACCUUGGAAAAUAUGUAAUAGGCUGACAGAAAUAUGAGCACGCCACUCCGGGAAAUUUAAGGAAA .....((((.(((((((((((.....)))))).........(((.(.(..((....))..).)............(((((.((......))..)).))).)))))))).))))....... ( -27.60, z-score = -0.85, R) >droYak2.chr2R 13650281 120 + 21139217 UGGAAAAAUCUCUUGACACGGGCUGACCGUGUGAAAUAUUUGAGAUGCUUGGGAAACGUUGGAAAAUAUCUAAUAGGCUGACAGGAAAGGUAGUACGCCACCCUGGAAAAUGCGAGGAAA ........((((((.((((((.....)))))).)).(((((.....((((.(....)((((((.....))))))))))...((((...(((.....)))..))))..))))).))))... ( -29.40, z-score = -1.27, R) >droEre2.scaffold_4845 6232122 120 + 22589142 UGAAAAAAUCGCUUGACACGGGCUGACCGUGUGAAAUAUUCGAGAUGCUUGGGAAACCUCGGAAAAUUUCUAAUAGGCUGACAGGAAAGUUAGUACGUCAGCAUGGUAAAUGUGAGGAAA ........((((((.((((((.....)))))).)).((((.(((((.((.((....))..))...))))).)))).((((((..............)))))).........))))..... ( -32.04, z-score = -2.03, R) >droAna3.scaffold_13266 16759948 115 - 19884421 ---GAAGAUCUCCAGACACGGGUUGACCGUGUGAAAUAUUCCAGAUGCUUGGGAAACCAUGGAAAAAAUUGAUUAGGAGAACAGAAAAACUCAUAGGUGAUUCGGAAAACUCGGAAAA-- ---...(.(((((..((((((.....))))))......(((((......(((....))))))))...........))))).)..................(((.((....)).)))..-- ( -29.60, z-score = -1.77, R) >dp4.chr3 5156355 110 + 19779522 ----------GCUUGACACGGGCUGACCGUGUGAAAUAUUCCAACUGUUAUGGCAGCACCCCAAUAUUGUCAACAGGCCUGCAGCAAAAGCAGUAGGCGAUGCCGGAAAAACGGUGGAAA ----------..((.((((((.....)))))).))...(((((.(((((.(((((.(.........(((....)))((((((.((....)).))))))).)))))....)))))))))). ( -39.70, z-score = -2.47, R) >droPer1.super_4 2983186 110 - 7162766 ----------GCUUGACACGGGCUGACCGUGUGAAAUAUUCCAACUGUUAUGGCAGCACCCCAAUAUUGUCAACAGGCCUGCAGCAAAAGCAGUAGGCAAUGCCGGAAAAACGGUGGAAA ----------..((.((((((.....)))))).))...(((((.(((((.(((((...........(((....)))((((((.((....)).))))))..)))))....)))))))))). ( -39.40, z-score = -2.47, R) >consensus UGAAAAGAUCUCUUGACACGGGCUGACCGUGUGAAAUAUUCGAGAUGCUUGGGAAACCUCGGAAAAUAUCUAAUAGGCUGACAGAAAAAGUAGUACGCCACUCCGGAAAAUCGGAGGAAA ...........((((((((((.....))))))..................((....))...............))))........................................... (-13.27 = -12.86 + -0.40)
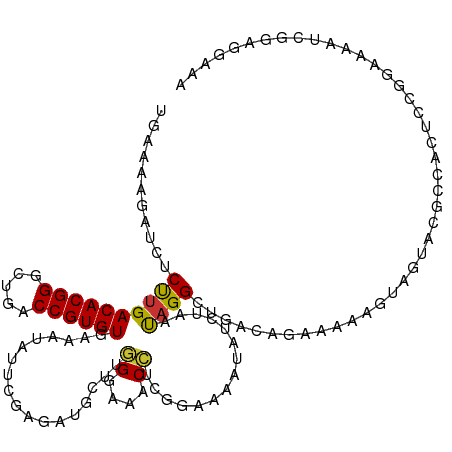
| Location | 9,458,292 – 9,458,400 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.86 |
| Shannon entropy | 0.54337 |
| G+C content | 0.45583 |
| Mean single sequence MFE | -31.19 |
| Consensus MFE | -13.27 |
| Energy contribution | -12.86 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.780230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9458292 108 - 21146708 ----AAUCGUGGUUGGUG--UAGUCCCAAAAUCAUGAAAAGAUCUCUUGACACGGGCUGACCGUGUGAAAUAUUAGAGAUGCUUGGGAAACCUCGGAAAAUAUCUAACAGGCUG ----....((.(((((((--(..(((............((((((((((.((((((.....)))))).)......)))))).)))((....))..)))..))).)))))..)).. ( -30.60, z-score = -2.09, R) >droSim1.chr2R 7901799 108 - 19596830 ----AAUCGUGGUUGGUG--UUGUCCCAAAACCAUGAAAAGAUCUCUUGACACGGGCUGACCGUGUGAAAUAUUCGAGAUGCUUGGGAAACCUUGGAAAAUAUCUAAUAGGCUG ----..((((((((((.(--....))))..)))))))....(((((((.((((((.....)))))).))......)))))((((((....))(((((.....))))).)))).. ( -33.10, z-score = -3.00, R) >droSec1.super_1 6976395 108 - 14215200 ----AAUCGUGGUUGGUG--CUGUCCCAAAACCAUGAAGAGAUCUCUUGACACGGGCUGACCGUGUGAAAUAUUCGAGAUGCUUGGGAAACCUUGGAAAAUAUGUAAUAGGCUG ----..((((((((((..--.....)))..))))))).((((((((((.((((((.....)))))).))......))))).)))((....))...................... ( -30.30, z-score = -1.81, R) >droYak2.chr2R 13650321 108 + 21139217 ----AAUCGUGUUUGGUG--UUGUCCCAAAACCAUGGAAAAAUCUCUUGACACGGGCUGACCGUGUGAAAUAUUUGAGAUGCUUGGGAAACGUUGGAAAAUAUCUAAUAGGCUG ----..((((((((((.(--....))))))..)))))....(((((((.((((((.....)))))).))......)))))((((.(....)((((((.....)))))))))).. ( -27.80, z-score = -1.63, R) >droEre2.scaffold_4845 6232162 108 + 22589142 ----AAUCGUGGUUGGUG--UUGUCCCAAAUCCAUGAAAAAAUCGCUUGACACGGGCUGACCGUGUGAAAUAUUCGAGAUGCUUGGGAAACCUCGGAAAAUUUCUAAUAGGCUG ----..((((((((((.(--....)))))..)))))).......(((((((((((.....)))))).........(((((.((.((....))..))...)))))...))))).. ( -32.20, z-score = -2.37, R) >droAna3.scaffold_13266 16759986 97 - 19884421 -----------AAAAGUG--C-GUCCAAAAGGC---CGAAGAUCUCCAGACACGGGUUGACCGUGUGAAAUAUUCCAGAUGCUUGGGAAACCAUGGAAAAAAUUGAUUAGGAGA -----------.......--(-(.((....)).---))....(((((..((((((.....))))))......(((((......(((....))))))))...........))))) ( -28.50, z-score = -2.37, R) >dp4.chr3 5156395 110 + 19779522 AACCGAUCCUUUUGUGUGGAUGGUUCUUGGGUC----GGUGCCAGCUUGACACGGGCUGACCGUGUGAAAUAUUCCAACUGUUAUGGCAGCACCCCAAUAUUGUCAACAGGCCU ((((.((((........)))))))).(((((..----(.(((((...(.((((((.....)))))).)((((.......)))).))))).)..)))))....(((....))).. ( -33.50, z-score = -0.15, R) >droPer1.super_4 2983226 110 - 7162766 AACCGAUCCUUUUGUGUGGAUGGUUCUUGGGUC----GGUGCCAGCUUGACACGGGCUGACCGUGUGAAAUAUUCCAACUGUUAUGGCAGCACCCCAAUAUUGUCAACAGGCCU ((((.((((........)))))))).(((((..----(.(((((...(.((((((.....)))))).)((((.......)))).))))).)..)))))....(((....))).. ( -33.50, z-score = -0.15, R) >consensus ____AAUCGUGGUUGGUG__UUGUCCCAAAACCAUGAAAAGAUCUCUUGACACGGGCUGACCGUGUGAAAUAUUCGAGAUGCUUGGGAAACCUCGGAAAAUAUCUAAUAGGCUG .............................................((((((((((.....))))))..................((....))...............))))... (-13.27 = -12.86 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:27 2011