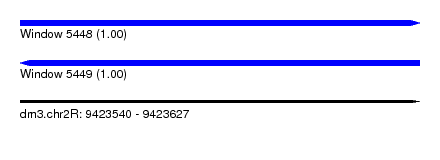
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,423,540 – 9,423,627 |
| Length | 87 |
| Max. P | 0.999453 |

| Location | 9,423,540 – 9,423,627 |
|---|---|
| Length | 87 |
| Sequences | 13 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 70.29 |
| Shannon entropy | 0.66208 |
| G+C content | 0.51890 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -25.65 |
| Energy contribution | -24.52 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
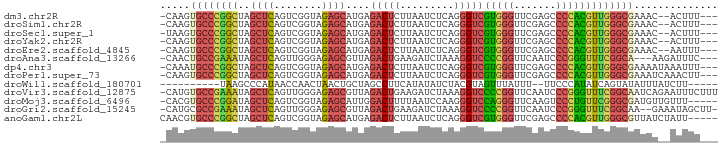
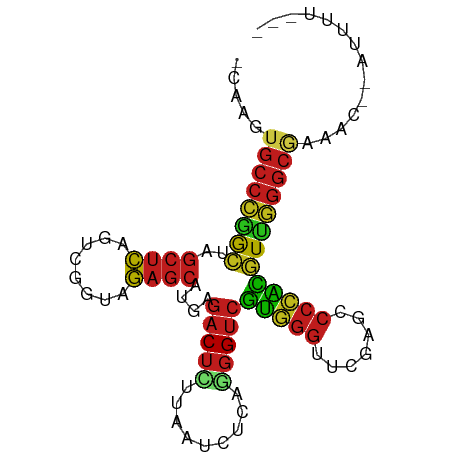
>dm3.chr2R 9423540 87 + 21146708 ---AAAGU--GUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCACUUG- ---.((((--(((..((....((((((.......))))))....((((((.....)))))).((((........))))..))..))))))).- ( -32.30, z-score = -2.84, R) >droSim1.chr2R 7867353 87 + 19596830 ---AAAGU--GUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCACUUG- ---.((((--(((..((....((((((.......))))))....((((((.....)))))).((((........))))..))..))))))).- ( -32.30, z-score = -2.84, R) >droSec1.super_1 6942371 87 + 14215200 ---AAAGU--GUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCACUUA- ---.((((--(((..((....((((((.......))))))....((((((.....)))))).((((........))))..))..))))))).- ( -32.30, z-score = -2.85, R) >droYak2.chr2R 13609723 87 - 21139217 ---AAAGU--GUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCACUUG- ---.((((--(((..((....((((((.......))))))....((((((.....)))))).((((........))))..))..))))))).- ( -32.30, z-score = -2.84, R) >droEre2.scaffold_4845 6196232 87 - 22589142 ---AAAUU--GUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCACUUG- ---.....--.....((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).....- ( -31.50, z-score = -3.19, R) >droAna3.scaffold_13266 13170229 86 - 19884421 ---GAAAUCUU---UGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCAGUUG- ---.......(---(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))))...- ( -30.90, z-score = -3.60, R) >dp4.chr3 17983371 89 + 19779522 ---AAAUUUAUUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCAUUUG- ---............((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).....- ( -31.50, z-score = -3.66, R) >droPer1.super_73 65986 89 + 267513 ---AAGUUUGAUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCACUUG- ---............((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).....- ( -31.50, z-score = -2.79, R) >droWil1.scaffold_180701 3763499 76 + 3904529 -----AAGAUAAAUAUACUGAUAUGGGAA--AAAUAAACUACGUAGAUAUAUGAAGGCUAGCAGUUAGUUGGUUAUGGGCUUA---------- -----.......((((((((...(((...--.......)))..))).)))))...(((((((.....))))))).........---------- ( -8.80, z-score = 0.64, R) >droVir3.scaffold_12875 9399628 92 - 20611582 AAAGAAAUUCUGAUUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCACAUG- ..............(((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))))....- ( -30.20, z-score = -3.49, R) >droMoj3.scaffold_6496 15505840 87 + 26866924 -----AAACAACAUCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCACGUG- -----..........(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).....- ( -29.40, z-score = -3.82, R) >droGri2.scaffold_15245 13524859 89 + 18325388 -AAGCUAUUUC--UUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCGCAUG- -..........--..((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))).....- ( -29.60, z-score = -3.05, R) >anoGam1.chr2L 7730467 88 - 48795086 -----AAUAGAUAACGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCACGUUG -----..........((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))...... ( -31.50, z-score = -3.22, R) >consensus ___AAAAU__GUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCACUUG_ ...............((((...(((((.......))))).....((((((.....)))))).((((........)))).....))))...... (-25.65 = -24.52 + -1.13)

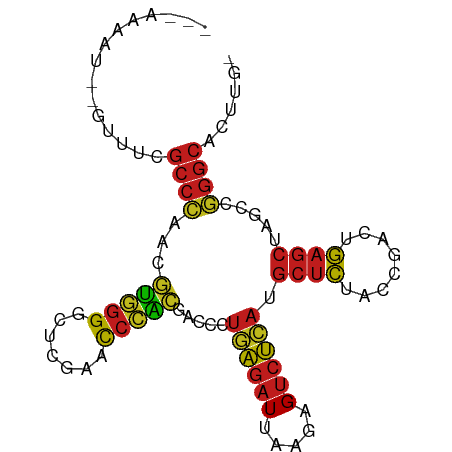
| Location | 9,423,540 – 9,423,627 |
|---|---|
| Length | 87 |
| Sequences | 13 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 70.29 |
| Shannon entropy | 0.66208 |
| G+C content | 0.51890 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -27.11 |
| Energy contribution | -25.82 |
| Covariance contribution | -1.29 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.996947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9423540 87 - 21146708 -CAAGUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAC--ACUUU--- -....((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))....--.....--- ( -32.90, z-score = -1.95, R) >droSim1.chr2R 7867353 87 - 19596830 -CAAGUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAC--ACUUU--- -....((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))....--.....--- ( -32.90, z-score = -1.95, R) >droSec1.super_1 6942371 87 - 14215200 -UAAGUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAC--ACUUU--- -....((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))....--.....--- ( -32.90, z-score = -2.00, R) >droYak2.chr2R 13609723 87 + 21139217 -CAAGUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAC--ACUUU--- -....((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))....--.....--- ( -32.90, z-score = -1.95, R) >droEre2.scaffold_4845 6196232 87 + 22589142 -CAAGUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAC--AAUUU--- -....((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))....--.....--- ( -32.90, z-score = -2.31, R) >droAna3.scaffold_13266 13170229 86 + 19884421 -CAACUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCA---AAGAUUUC--- -....((((((((..((((........))))...........((((....))))(((((.......)))))))))))))---........--- ( -27.50, z-score = -1.74, R) >dp4.chr3 17983371 89 - 19779522 -CAAAUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAAUAAAUUU--- -....((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))...........--- ( -33.00, z-score = -2.80, R) >droPer1.super_73 65986 89 - 267513 -CAAGUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAUCAAACUU--- -....((((((((..((((........))))....((((((.......))))))(((((.......)))))))))))))...........--- ( -32.90, z-score = -2.11, R) >droWil1.scaffold_180701 3763499 76 - 3904529 ----------UAAGCCCAUAACCAACUAACUGCUAGCCUUCAUAUAUCUACGUAGUUUAUUU--UUCCCAUAUCAGUAUAUUUAUCUU----- ----------.................(((((((((...........))).)))))).....--........................----- ( -4.10, z-score = 0.03, R) >droVir3.scaffold_12875 9399628 92 + 20611582 -CAUGUGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAAUCAGAAUUUCUUU -....((((((((..((((........))))...........((((....))))(((((.......))))))))))))).............. ( -27.60, z-score = -1.38, R) >droMoj3.scaffold_6496 15505840 87 - 26866924 -CACGUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGAUGUUGUUU----- -.(((((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))).)))).....----- ( -34.80, z-score = -3.44, R) >droGri2.scaffold_15245 13524859 89 - 18325388 -CAUGCGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAA--GAAAUAGCUU- -.....(((((((..((((........))))...........((((....))))(((((.......))))))))))))..--..........- ( -27.00, z-score = -1.07, R) >anoGam1.chr2L 7730467 88 + 48795086 CAACGUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGUUAUCUAUU----- ......(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))..........----- ( -32.70, z-score = -1.96, R) >consensus _CAAGUGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAC__AUUUU___ ......(((((((..((((........))))....(((((.........)))))(((((.......))))))))))))............... (-27.11 = -25.82 + -1.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:23 2011