| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,412,832 – 9,412,926 |
| Length | 94 |
| Max. P | 0.984962 |

| Location | 9,412,832 – 9,412,926 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 56.95 |
| Shannon entropy | 0.85417 |
| G+C content | 0.56092 |
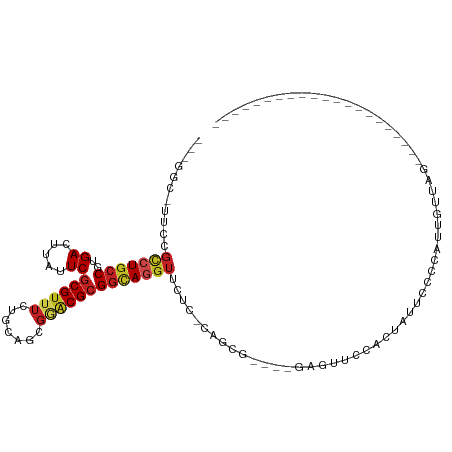
| Mean single sequence MFE | -30.24 |
| Consensus MFE | -13.77 |
| Energy contribution | -14.18 |
| Covariance contribution | 0.41 |
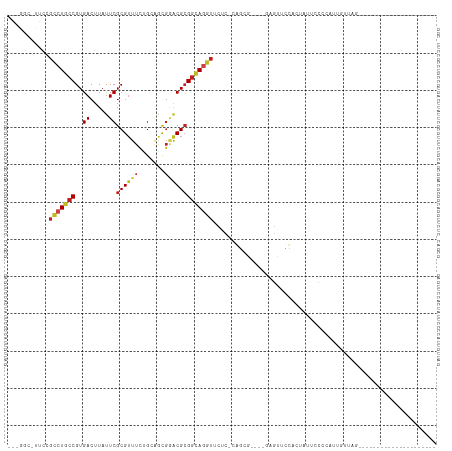
| Combinations/Pair | 1.27 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
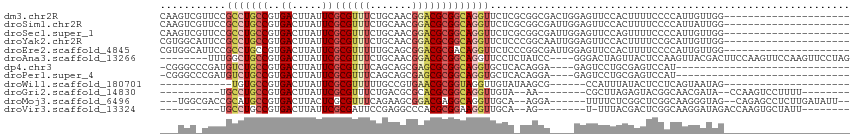
>dm3.chr2R 9412832 94 + 21146708 CAAGUCGUUCCGCCUGCCGUGACUUAUUCGCGUUUCUGCAACGGACGCGGCAGGUUCUCGCGGCGACUGGAGUUCCACUUUUCCCCAUUGUUGG--------------------- ..((((((..(((((((((((......((.((((.....)))))))))))))))....))..))))))((((........))))(((....)))--------------------- ( -32.10, z-score = -1.27, R) >droSim1.chr2R 7854924 94 + 19596830 CAAGUCGUUCCGCCUGCCGUGACUUAUUCGCGUUUCUGCAACGGACGCGGCAGGUUCUCGCGGCGAUUGGAGUUCCACUUUUCCCCAUUAUUGG--------------------- ...(((((...((((((((((......((.((((.....))))))))))))))))....)))))....((((........))))(((....)))--------------------- ( -30.50, z-score = -1.27, R) >droSec1.super_1 6931692 94 + 14215200 CAAGUCGUUCCGCCUGCCGUGACUUAUUCGCGUUUCUGCAACGGACGCGGCAGGUUCUCGCGGCGAUUGGAGUUCCAGUUUUCCCCAUUGUUGG--------------------- ...(((((...((((((((((......((.((((.....))))))))))))))))....)))))((((((....))))))....(((....)))--------------------- ( -33.00, z-score = -1.54, R) >droYak2.chr2R 13599228 94 - 21139217 CGUGGCAUUCCGCCUGCCGUGACUUAUUCGCGUUUCUGCAACGGACGCGGCAGGUUCUCCCGGCAAUUGGAGUUCCACUUUUCCGCAUUGUUGG--------------------- ...........((((((((((......((.((((.....))))))))))))))))....(((((((((((((........))))).))))))))--------------------- ( -32.70, z-score = -1.45, R) >droEre2.scaffold_4845 6185732 94 - 22589142 CGUGGCAUUCCGCCUGCCGUGACUUAUUCGCGUUUUUGCAGCGGACGCGACAGGUUCUCCCGGCGAUUGGAGUUCCACUUUUCCCCAUUGUUGG--------------------- .((((.((((((..(((((.(((((..((((((((.......)))))))).)))))....)))))..)))))).))))......(((....)))--------------------- ( -33.80, z-score = -2.09, R) >droAna3.scaffold_13266 16711741 103 + 19884421 --------UUUGGCUGCCGUGACUUAUUCGCGUUUCUGCAACGGACGCGGCAGGUUCCUCUAUCC----GGGACUAGUUACUCCAAGUUACGACUUCCAAGUUCCAAGUUCCUAG --------(((((..(.((((((((....(((....)))...(((.(..((.((((((.......----)))))).))..)))))))))))).)..))))).............. ( -30.00, z-score = -1.43, R) >dp4.chr3 8251605 81 + 19779522 -CGGGCCCGAUGUCUGCCGUGACUUAUUCGCGUUUCAGCAGCGAGCGCGGCAGGUGCUCACAGGA----GAGUCCUGCGAGUCCAU----------------------------- -.((((.((.(..((((((((.((..((.((......))))..))))))))))..)....((((.----....)))))).))))..----------------------------- ( -33.80, z-score = -1.38, R) >droPer1.super_4 2132577 81 + 7162766 -CGGGCCCGAUGUCUGCCGUGACUUAUUCGCGUUUCAGCAGCGAGCGCGGCAGGUGCUCACAGGA----GAGUCCUGCGAGUCCAU----------------------------- -.((((.((.(..((((((((.((..((.((......))))..))))))))))..)....((((.----....)))))).))))..----------------------------- ( -33.80, z-score = -1.38, R) >droWil1.scaffold_180701 3746124 76 + 3904529 ------------UGUGCCGUGACUUAUUCGCGUUUUUGCCGUGAACGCGGUAGGUUGUAUAAGCG------CCAUUUAUACUCCUCAGUAAUAG--------------------- ------------.((((.(..((((((.(((((((.......)))))))))))))..)....)))------)......((((....))))....--------------------- ( -20.10, z-score = -1.17, R) >droGri2.scaffold_14830 4778659 85 - 6267026 ----------UGCCUGCCGUGACUUAUUCGCGUUUCUGACGCGCACGCGGCAGGUUGUA--AA--------CGCUUAGAGUACGGCAACGAUA--CCAAGUCCUUUU-------- ----------.((((((((((.......((((((...))))))..))))))))))....--..--------.((((.(.(((((....)).))--))))))......-------- ( -30.20, z-score = -2.02, R) >droMoj3.scaffold_6496 606829 100 - 26866924 ---UGGCGACCGCAUGCCGUGACUUACUCGCGUUUCAGAAGCGGACGAGGCAGGUUGCA--AGGA------UUUUCUCGGCUCGGCAAGGGUAG--CAGAGCCUCUUGAUAUU-- ---..((((((...((((((((.....))))((((.......))))..)))))))))).--....------.......(((((.((.......)--).)))))..........-- ( -30.10, z-score = 0.83, R) >droVir3.scaffold_13324 2611728 86 + 2960039 ----------UGCCUGCCGUGACUUAUUCGCGAUUCCGAGGCCCACGCGGAAGGUUGCA--AG--------U-UUUACGACUCGGCAAGGAUAGACCAAGUGCUAUU-------- ----------..(((((((..((((.((((((...(....)....))))))))))..).--((--------(-(....)))).))).)))((((........)))).-------- ( -22.80, z-score = 0.30, R) >consensus ___GGC_UUCCGCCUGCCGUGACUUAUUCGCGUUUCUGCAGCGGACGCGGCAGGUUCUC_CAGCG____GAGUUCCACUAUUCCCCAUUGUUAG_____________________ ...........(((((((..((.....))((((((.......)))))))))))))............................................................ (-13.77 = -14.18 + 0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:21 2011