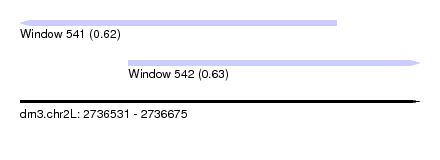
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,736,531 – 2,736,675 |
| Length | 144 |
| Max. P | 0.628126 |

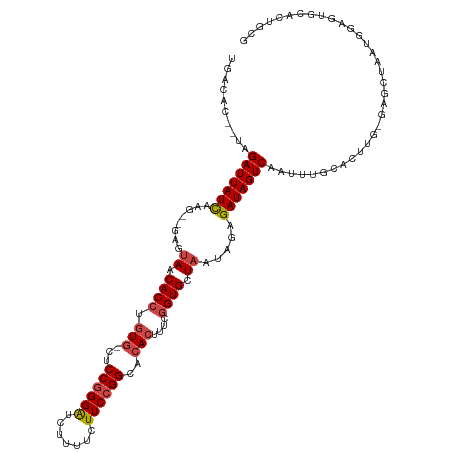
| Location | 2,736,531 – 2,736,645 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.55 |
| Shannon entropy | 0.45031 |
| G+C content | 0.46730 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -16.73 |
| Energy contribution | -16.98 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.615712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2736531 114 - 23011544 UGACAC--UAGAUUAUCAAG--GAGUAACACCUGUG-CUCCGGGAUCUUUUCUUCCGGCACACUUUCGGUGCUAAUAGAGAUAGUCAAUUCGCACUUG-GAGCUAAUGGAGUGCACUGCG ......--..(((((((..(--(((((.(....)))-)))).(((........)))(((((.......)))))......))))))).....((((((.-(......).))))))...... ( -32.40, z-score = -1.09, R) >droAna3.scaffold_12984 200665 115 - 754457 UGGCUUUAUAGAUUAUUAAAAUGAGUAUCACCUGUGACACCAGGGUCUUUUCUUCCGGUGAAGCUCGAGUGUU-----UGAUAGUCUAUUUCCAGCAGAAAGCUAAUUCAGUUAUUUUUA ((((((((((((((((((((.(((((.(((((..((....))(((........)))))))).)))))....))-----))))))))))).........)))))))............... ( -31.00, z-score = -2.52, R) >droEre2.scaffold_4929 2789094 95 - 26641161 UGACUC--CGGAUUAUCAAG--GAGUAGCACCUGUG-CUCCGGGAUCUUCUCUUCCGGCCCACUUUCGGUGCUAAUAGAGAUAGUCCAUGGAGAACUG-CG------------------- ...(((--(((((((((...--...(((((((.(((-..((((((.......))))))..)))....))))))).....))))))))..)))).....-..------------------- ( -37.70, z-score = -3.21, R) >droYak2.chr2L 2731617 114 - 22324452 UGACUC--UGGAUUAUCAAG--GAGUAGCACCUGUG-CUCCGGGAUCUUUUCUUCCGGCACACUUUCGGUGCUAAUAGAGAUAGUCAAUUUGCACUGG-CAGCCAAUGGAGUGCACUGCG ((((((--(.(((((((...--...(((((((.(((-..((((((.......))))))..)))....))))))).....)))))))...((((....)-))).....))))).))..... ( -36.90, z-score = -1.28, R) >droSec1.super_5 902673 114 - 5866729 UGAGAC--UAGAUUAUCAAG--GAGUAACACCUGUG-CUCCGGGAUCUUUUCUUCCGGCACACUUUCGGUGCUAAUAGAGAUAGUCAAUUUCCACUUG-GAGCUAAUGGAGUGCACUGGG .....(--(((.......((--(.......)))(..-(((((.(((..(((((...(((((.......)))))...)))))..)))...((((....)-)))....)))))..).)))). ( -32.90, z-score = -0.88, R) >droSim1.chr2L 2696929 114 - 22036055 UGACAC--UAGAUUAUCAAG--GAGUAACACCUGUG-CUCCGGGAUCUUUUCUUCCGGCACACUUUCGGUGCUAAUAGAGAUAGUCAAUUCGCACUUG-GAGCUAAUGGAGUGCACUGCG ......--..(((((((..(--(((((.(....)))-)))).(((........)))(((((.......)))))......))))))).....((((((.-(......).))))))...... ( -32.40, z-score = -1.09, R) >consensus UGACAC__UAGAUUAUCAAG__GAGUAACACCUGUG_CUCCGGGAUCUUUUCUUCCGGCACACUUUCGGUGCUAAUAGAGAUAGUCAAUUUGCACUUG_GAGCUAAUGGAGUGCACUGCG ..........(((((((......((...((((.(((...((((((.......))))))..)))....))))))......))))))).................................. (-16.73 = -16.98 + 0.25)


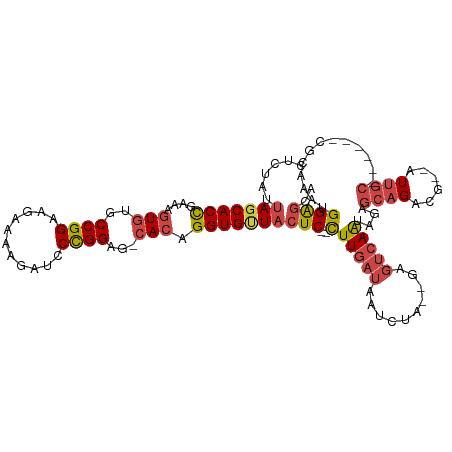
| Location | 2,736,570 – 2,736,675 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.73 |
| Shannon entropy | 0.36501 |
| G+C content | 0.48069 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -16.83 |
| Energy contribution | -18.58 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.628126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2736570 105 + 23011544 CUCUAUUAGCACCGAAAGUGUGCCGGAAGAAAAGAUCCCGGAG-CACAGGUGUUACUC--CUUGAUAAUCUA--GUGUCAAUAGAGCAGACG--AUUGC------CGCAAAUGGGGCA (((((((((((((....((((.((((...........)))).)-))).)))))))...--..(((((.....--.)))))))))))......--..(((------(.(....).)))) ( -33.00, z-score = -2.09, R) >droAna3.scaffold_12984 200705 105 + 754457 -----CAAACACUCGAGCUUCACCGGAAGAAAAGACCCUGGUGUCACAGGUGAUACUCAUUUUAAUAAUCUAUAAAGCCAGCAGAUCAGGGU--GUUAU------CUCUAAUGGAAAA -----.............((((..(((.((.((.((((((((((((....))))))...........((((...........))))))))))--.)).)------))))..))))... ( -22.80, z-score = -0.60, R) >droEre2.scaffold_4929 2789114 113 + 26641161 CUCUAUUAGCACCGAAAGUGGGCCGGAAGAGAAGAUCCCGGAG-CACAGGUGCUACUC--CUUGAUAAUCCG--GAGUCAAUAGAGCAGCCGCAAUAGCGAUUGCCGAAAAUGGAGCA (((((((((((((....(((..((((..((.....))))))..-))).))))))((((--(..........)--)))).....(.(((..(((....)))..))))...))))))).. ( -38.30, z-score = -2.36, R) >droYak2.chr2L 2731656 107 + 22324452 CUCUAUUAGCACCGAAAGUGUGCCGGAAGAAAAGAUCCCGGAG-CACAGGUGCUACUC--CUUGAUAAUCCA--GAGUCAAUAGAGCAGACG--AUUGC----CCCAAAAACGGAGCA (((((((((((((....((((.((((...........)))).)-))).))))))((((--..((......))--)))).)))))))......--..(((----.((......)).))) ( -32.80, z-score = -3.10, R) >droSec1.super_5 902712 105 + 5866729 CUCUAUUAGCACCGAAAGUGUGCCGGAAGAAAAGAUCCCGGAG-CACAGGUGUUACUC--CUUGAUAAUCUA--GUCUCAAUAGAGCAGACG--AUUGC------CGCAAAUGGGGCA (((((((((((((....((((.((((...........)))).)-))).))))))....--...(((......--)))..)))))))......--..(((------(.(....).)))) ( -31.20, z-score = -1.49, R) >droSim1.chr2L 2696968 105 + 22036055 CUCUAUUAGCACCGAAAGUGUGCCGGAAGAAAAGAUCCCGGAG-CACAGGUGUUACUC--CUUGAUAAUCUA--GUGUCAAUAGAGCAGACG--AUUGC------CGCAAAUGGGGCA (((((((((((((....((((.((((...........)))).)-))).)))))))...--..(((((.....--.)))))))))))......--..(((------(.(....).)))) ( -33.00, z-score = -2.09, R) >consensus CUCUAUUAGCACCGAAAGUGUGCCGGAAGAAAAGAUCCCGGAG_CACAGGUGUUACUC__CUUGAUAAUCUA__GAGUCAAUAGAGCAGACG__AUUGC______CGCAAAUGGAGCA (((((((((((((....(((..((((...........))))...))).)))))).(((...(((((..........)))))..))).......................))))))).. (-16.83 = -18.58 + 1.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:12:11 2011