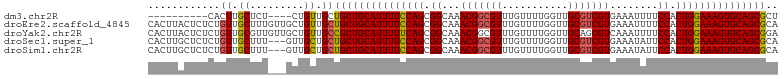
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,370,228 – 9,370,319 |
| Length | 91 |
| Max. P | 0.993763 |

| Location | 9,370,228 – 9,370,319 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 87.70 |
| Shannon entropy | 0.21436 |
| G+C content | 0.49729 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -27.42 |
| Energy contribution | -27.66 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
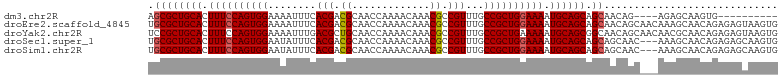
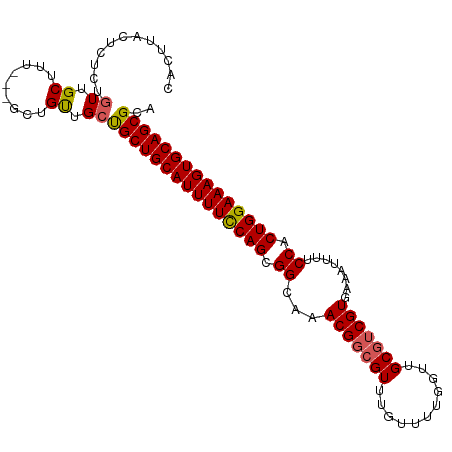
>dm3.chr2R 9370228 91 + 21146708 AGCGCUGCACUUUCCAGUGGAAAAUUUCACGACGCAACCAAAACAAACGCCGUUUGCCGCUGGAAAAUGCAGCAGCAACAG----AGAGCAAGUG---------- .((((((((.((((((((((........(((.((.............)).)))...)))))))))).)))))).)).....----..........---------- ( -30.42, z-score = -2.49, R) >droEre2.scaffold_4845 6143405 105 - 22589142 UGCGCUGCACUUUCCAGUGGAAAAUUUCACGACGCAACCAAAACAAACGCCGUUUGCCGCUGGAAAAUGCAGCAGCAACAGCAACAAAGCAACAGAGAGUAAGUG (((((((((.((((((((((........(((.((.............)).)))...)))))))))).)))))).((....))......))).............. ( -32.82, z-score = -2.43, R) >droYak2.chr2R 13555239 105 - 21139217 UCCGCUGCACUUUCCAGUGGAAAAUUUGACGCUGCAACCAAAACAAACGCCGUUUGCCGCUGAAAAAUGCAGCGGCAACAGCAACAACGCAACAGAGAGUAAGUG ..((((..(((((((((((..........)))))..............((.(((.(((((((.......)))))))))).))............)))))).)))) ( -32.40, z-score = -2.07, R) >droSec1.super_1 6889504 102 + 14215200 UGCGCUGCACUUUCCAGUGGAAUAUUUCACGACGCAACCAAAACAAACGCCGUUUGCCGCUGGAAAAUGCAGCAGCAGCAAC---AAAGCAACAGAGAGCAAGUG (((((((((.((((((((((........(((.((.............)).)))...)))))))))).))))))....((...---...))........))).... ( -33.02, z-score = -2.37, R) >droSim1.chr2R 7820833 102 + 19596830 UGCGCUGCACUUUCCAGUGGAAUAUUUCACGACGCAACCAAAACAAACGCCGUUUGCCGCUGGAAAAUGCAGCAGCAGCAAC---AAAGCAACAGAGAGCAAGUG (((((((((.((((((((((........(((.((.............)).)))...)))))))))).))))))....((...---...))........))).... ( -33.02, z-score = -2.37, R) >consensus UGCGCUGCACUUUCCAGUGGAAAAUUUCACGACGCAACCAAAACAAACGCCGUUUGCCGCUGGAAAAUGCAGCAGCAACAGC___AAAGCAACAGAGAGCAAGUG .((((((((.((((((((((........(((.((.............)).)))...)))))))))).)))))).))............................. (-27.42 = -27.66 + 0.24)
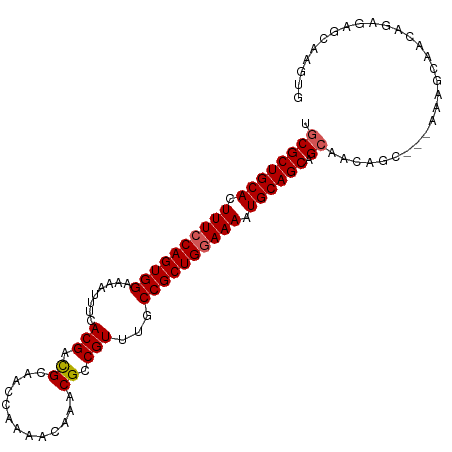
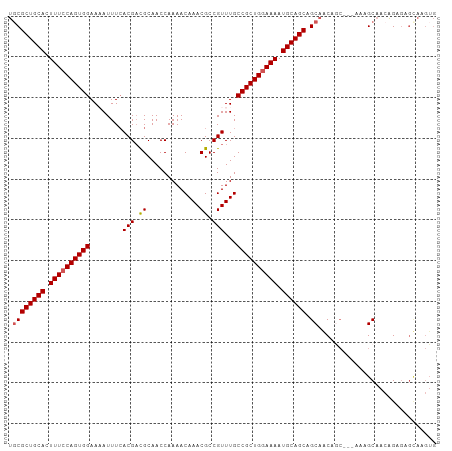
| Location | 9,370,228 – 9,370,319 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 87.70 |
| Shannon entropy | 0.21436 |
| G+C content | 0.49729 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -33.46 |
| Energy contribution | -33.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9370228 91 - 21146708 ----------CACUUGCUCU----CUGUUGCUGCUGCAUUUUCCAGCGGCAAACGGCGUUUGUUUUGGUUGCGUCGUGAAAUUUUCCACUGGAAAGUGCAGCGCU ----------..........----.....((.((((((((((((((.((...(((((((...........)))))))........)).)))))))))))))))). ( -36.70, z-score = -3.36, R) >droEre2.scaffold_4845 6143405 105 + 22589142 CACUUACUCUCUGUUGCUUUGUUGCUGUUGCUGCUGCAUUUUCCAGCGGCAAACGGCGUUUGUUUUGGUUGCGUCGUGAAAUUUUCCACUGGAAAGUGCAGCGCA ...............((......))...(((.((((((((((((((.((...(((((((...........)))))))........)).))))))))))))))))) ( -38.50, z-score = -2.66, R) >droYak2.chr2R 13555239 105 + 21139217 CACUUACUCUCUGUUGCGUUGUUGCUGUUGCCGCUGCAUUUUUCAGCGGCAAACGGCGUUUGUUUUGGUUGCAGCGUCAAAUUUUCCACUGGAAAGUGCAGCGGA .........(((((((((((((((((((((((((((.......))))))).)))))))............))))).....(((((((...))))))))))))))) ( -40.10, z-score = -2.63, R) >droSec1.super_1 6889504 102 - 14215200 CACUUGCUCUCUGUUGCUUU---GUUGCUGCUGCUGCAUUUUCCAGCGGCAAACGGCGUUUGUUUUGGUUGCGUCGUGAAAUAUUCCACUGGAAAGUGCAGCGCA ....(((.....((.((...---...)).)).((((((((((((((.((...(((((((...........)))))))........)).))))))))))))))))) ( -39.90, z-score = -2.90, R) >droSim1.chr2R 7820833 102 - 19596830 CACUUGCUCUCUGUUGCUUU---GUUGCUGCUGCUGCAUUUUCCAGCGGCAAACGGCGUUUGUUUUGGUUGCGUCGUGAAAUAUUCCACUGGAAAGUGCAGCGCA ....(((.....((.((...---...)).)).((((((((((((((.((...(((((((...........)))))))........)).))))))))))))))))) ( -39.90, z-score = -2.90, R) >consensus CACUUACUCUCUGUUGCUUU___GCUGUUGCUGCUGCAUUUUCCAGCGGCAAACGGCGUUUGUUUUGGUUGCGUCGUGAAAUUUUCCACUGGAAAGUGCAGCGCA .............................((.((((((((((((((.((...(((((((...........)))))))........)).)))))))))))))))). (-33.46 = -33.90 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:15 2011