
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,368,504 – 9,368,610 |
| Length | 106 |
| Max. P | 0.788489 |

| Location | 9,368,504 – 9,368,610 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.30 |
| Shannon entropy | 0.45929 |
| G+C content | 0.37705 |
| Mean single sequence MFE | -14.28 |
| Consensus MFE | -8.03 |
| Energy contribution | -8.03 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
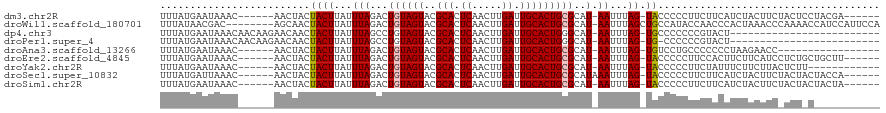
>dm3.chr2R 9368504 106 - 21146708 UUUAUGAAUAAAC------AACUACUACUUAUUUAGACUGUAGUACGCACUCAACUUGAUUGCACUGCGCAU-AAUUUAG-UACCCCCUUCUUCAUCUACUUCUACUCCUACGA------ ...(((((.....------...(((((.....(((...((((((..(((.((.....)).)))))))))..)-))..)))-))........)))))..................------ ( -12.29, z-score = -1.31, R) >droWil1.scaffold_180701 1100202 111 + 3904529 UUUAUAACGAC--------AGCAACUACUUAUUUAGACUGUAGUACGCACUCAACUUGAUUGCACUGCGCAU-AAUUUAGCUGCCAUACCAACCCACUAAACCCAAAACCAUCCAUUCCA ........(.(--------(((..(((......)))...(((((..(((.((.....)).))))))))....-......)))).)................................... ( -14.90, z-score = -1.91, R) >dp4.chr3 8201666 93 - 19779522 UUUAUGAAUAAACAACAAGAACAACUACUUAUUUAGCCUGUAGUACGCACUCAACUUGAUUGCACUGGGCAU-AAUUUAG-UGCCCCCCCGUACU------------------------- ....(((((((.................)))))))......((((((((.((.....)).)))...((((((-......)-)))))....)))))------------------------- ( -19.23, z-score = -2.04, R) >droPer1.super_4 2081880 92 - 7162766 UUUAUGAAUAAACAACAAGAACAACUACUUAUUUAGCCUGUAGUACGCACUCAACUUGAUUGCACUGGGCAU-AAUUUAG-UG-CCCCCCGUACU------------------------- ....(((((((.................)))))))......((((((((.((.....)).)))...((((((-......)-))-)))...)))))------------------------- ( -19.23, z-score = -1.98, R) >droAna3.scaffold_13266 16668411 95 - 19884421 UUUAUGAAUAAAC------AACUACUACUUAUUUAGACUGUAGUACGCACUCAACUUGAUUGCACUGCGCAU-AAUUUAG-UGUCCUGCCCCCCCUAAGAACC----------------- .............------............(((((...(((((..(((.((.....)).))))))))(((.-.((....-.))..))).....)))))....----------------- ( -12.10, z-score = -0.54, R) >droEre2.scaffold_4845 6141501 106 + 22589142 UUUAUGAAUAAAC------AACUACUACUUAUUUAGACUGUAGUACGCACUCAACUUGAUUGCACUGCGCAU-AAUUUAG-UACCCCCUUCCACUUCUUCAUCCUCUGCUGCUU------ .............------......((((...(((...((((((..(((.((.....)).)))))))))..)-))...))-))...............................------ ( -11.60, z-score = -0.45, R) >droYak2.chr2R 13550611 100 + 21139217 UUUAUGAAUAAAC------AACUACUACUUAUUUAGACUGUAGUACGCACUCAACUUGAUUGCACUGCGCAU-AAUUUAG-UACCCCCUUCUAUUUCUUCUUACUCUU------------ .....(((.....------......((((...(((...((((((..(((.((.....)).)))))))))..)-))...))-))...........)))...........------------ ( -11.95, z-score = -1.56, R) >droSec1.super_10832 153 107 - 1332 UUUAUGAUUAAAC------AACUACUACUUAUUUAGACUGUAGUACGCACUCAACUUGAUUGCACUGCGCAUAAAUUUAG-UACCCCCUUCUUCAUCUACUUCUACUACUACCA------ ...((((......------...(((((...(((((...((((((..(((.((.....)).)))))))))..))))).)))-)).........))))..................------ ( -14.97, z-score = -2.81, R) >droSim1.chr2R 7817700 106 - 19596830 UUUAUGAAUAAAC------AACUACUACUUAUUUAGACUGUAGUACGCACUCAACUUGAUUGCACUGCGCAU-AAUUUAG-UACCCCCUUCUUCAUCUACUUCUACUACUACUA------ ...(((((.....------...(((((.....(((...((((((..(((.((.....)).)))))))))..)-))..)))-))........)))))..................------ ( -12.29, z-score = -1.41, R) >consensus UUUAUGAAUAAAC______AACUACUACUUAUUUAGACUGUAGUACGCACUCAACUUGAUUGCACUGCGCAU_AAUUUAG_UACCCCCUUCUACUUCUACUUC_ACU_C___________ .......................(((((...........)))))..(((.((.....)).)))......................................................... ( -8.03 = -8.03 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:13 2011