
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,348,060 – 9,348,140 |
| Length | 80 |
| Max. P | 0.560092 |

| Location | 9,348,060 – 9,348,140 |
|---|---|
| Length | 80 |
| Sequences | 8 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 74.70 |
| Shannon entropy | 0.47939 |
| G+C content | 0.57195 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -13.92 |
| Energy contribution | -13.54 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.560092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
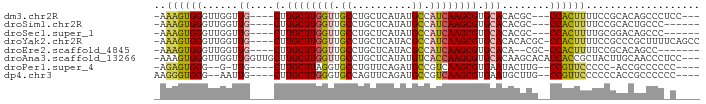
>dm3.chr2R 9348060 80 + 21146708 -AAAGUGGGUUGGUUG----CUUGCUUGGUUGCCUGCUCAUAUGCCAUCAAGCGUGCACACGC---CCACUUUUCCGCACAGCCCUCC--- -(((((((((.(..((----(.((((((((.((.((....)).)).)))))))).)))..)))---)))))))...............--- ( -27.60, z-score = -1.58, R) >droSim1.chr2R 7796307 77 + 19596830 -AAAGUGGGUUGGUUG----CUUGCUUGGUUGCCUGCUCAUAUGCCAUCAAGCGUGCACACGC---CCACUUUUCCGCACUGCCC------ -(((((((((.(..((----(.((((((((.((.((....)).)).)))))))).)))..)))---)))))))............------ ( -27.60, z-score = -1.64, R) >droSec1.super_1 6867119 77 + 14215200 -AAAGUGGGUUGGUUG----CUUGCUUGGUUGCCUGCUCAUAUGCCAUCAAGCGUGCACACGC---CCACUUUUGCGGACAGCCC------ -(((((((((.(..((----(.((((((((.((.((....)).)).)))))))).)))..)))---))))))).((.....))..------ ( -28.40, z-score = -1.11, R) >droYak2.chr2R 13529612 85 - 21139217 -AAAGUGGGUUGGUUG----CUUGCUUGGUUGCCUGCUCAUACGCCAUCAAGCGUGCACACACGC-CCACUUUUCCGCCCGCUUUUCAGCC -(((((((((((..((----(.((((((((.((..........)).)))))))).)))..)).))-))))))).......(((....))). ( -30.00, z-score = -2.05, R) >droEre2.scaffold_4845 6120328 76 - 22589142 -AAAGUGGGUUGGUUG----CUUGCUUGGUUGCCUGCUCAUACGCCAUCAAGCGUGCACA--CGC-CCACUUUUCCGCACAGCC------- -(((((((((.(..((----(.((((((((.((..........)).)))))))).)))..--)))-)))))))...........------- ( -27.60, z-score = -1.60, R) >droAna3.scaffold_13266 16650435 87 + 19884421 -AAAGUGGGUUGGUUGGUUGCUUGCUUGGUUGCCUGCUCAUAUGUCACCAAGCGUGCACAAGCACACCACCGCUACUUGCAACCCUCC--- -...(.((((((((.(((.....(((((((.((.((....)).)).)))))))((((....))))))))))((.....))))))).).--- ( -31.50, z-score = -1.58, R) >droPer1.super_4 2062570 76 + 7162766 -AGAGUGGG--G-UUG----CUUGCUUAGGUGCCUGUUCAGAUGCCGUCAAGCGUGAAUACUUG--CCGUUCCCCC-ACCGCCCCCC---- -.(.(((((--(-..(----(..((((.(((..((....))..)))...))))(..(....)..--).))..))))-))).......---- ( -23.90, z-score = -1.41, R) >dp4.chr3 8184421 79 + 19779522 AAGGGUGGG--AAUUG----CUUGCUUGGGUGCCAGUUCAGAUGCCGUCAAGCGUGAAUGCUUG--CCGUUCCCCCCACCGCCCCCC---- ..(((.(((--(((.(----(..((((((((..(......)..)))..)))))((....))..)--).)))))))))..........---- ( -26.30, z-score = -0.48, R) >consensus _AAAGUGGGUUGGUUG____CUUGCUUGGUUGCCUGCUCAUAUGCCAUCAAGCGUGCACACGCG__CCACUUUUCCGCACAGCCC_C____ ..((((((..............((((((((.((..........)).))))))))............))))))................... (-13.92 = -13.54 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:05 2011