
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,325,368 – 9,325,445 |
| Length | 77 |
| Max. P | 0.990894 |

| Location | 9,325,368 – 9,325,445 |
|---|---|
| Length | 77 |
| Sequences | 14 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 83.82 |
| Shannon entropy | 0.36292 |
| G+C content | 0.58643 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -26.67 |
| Energy contribution | -24.38 |
| Covariance contribution | -2.29 |
| Combinations/Pair | 1.90 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9325368 77 + 21146708 GCGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAA----- ..(((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......)))))))..----- ( -28.50, z-score = -1.35, R) >droGri2.scaffold_15112 1241212 81 - 5172618 UCGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCCCAUGGGCCACAUCA- ..(((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....)))))......- ( -28.90, z-score = -1.11, R) >droMoj3.scaffold_6496 13663476 76 + 26866924 AUGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCCCAUGGGCCA------ .((((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....))))))------ ( -28.80, z-score = -1.18, R) >droVir3.scaffold_12875 1837644 82 + 20611582 GUGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAA-------CCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGCAGCA .((((((((.(((((.........)))).((((((..((((-------((....))))))..).)))))....).))))))))...... ( -37.10, z-score = -1.53, R) >droWil1.scaffold_180772 6390000 77 + 8906247 AAGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAU----- ..(((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......)))))))..----- ( -28.40, z-score = -1.87, R) >dp4.chr3 6230985 78 - 19779522 GUGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGA---- .((((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))))..---- ( -29.10, z-score = -1.55, R) >droPer1.super_2 8071538 78 + 9036312 GUGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGA---- .((((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))))..---- ( -29.10, z-score = -1.55, R) >droAna3.scaffold_13266 16626120 77 + 19884421 ACGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAG----- ..(((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....)))))..----- ( -28.90, z-score = -1.41, R) >droEre2.scaffold_4845 6097252 77 - 22589142 GCGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAA----- ..(((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....)))))..----- ( -28.90, z-score = -1.09, R) >droYak2.chr2R 13505628 77 - 21139217 GCGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAA----- ..(((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....)))))..----- ( -28.90, z-score = -1.09, R) >droSec1.super_1 6844491 77 + 14215200 GCGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAA----- ..(((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....)))))..----- ( -28.90, z-score = -1.09, R) >droSim1.chr2R 7772970 77 + 19596830 ACGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAA----- ..(((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....)))))..----- ( -28.90, z-score = -1.36, R) >triCas2.chrUn_43 98017 83 - 275627 -CGGUUCGUUAGCUCAGGUGGUUAGAGCACCGCACUAAGCAGCAGCCUUGCGGAGGUCGUAGGUUCGAAUCCUACAUGAGCCAA----- -.((((((((.(((.(((((((......))))).)).)))))).(((((...))))).(((((.......)))))..)))))..----- ( -29.80, z-score = -1.93, R) >anoGam1.chr2R 13948018 74 - 62725911 --GGCUCCAUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGUGAGUUCGAUCCUCUCUGGAGCCA------ --(((((((((((...))))...((((.((((.(((....(-------(((....).))).))).)))).)))).))))))).------ ( -30.50, z-score = -2.76, R) >consensus GCGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA_______CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAA_____ ..(((((((..((((.........)))).(((((..............))))).....(.(((.......))).))))))))....... (-26.67 = -24.38 + -2.29)

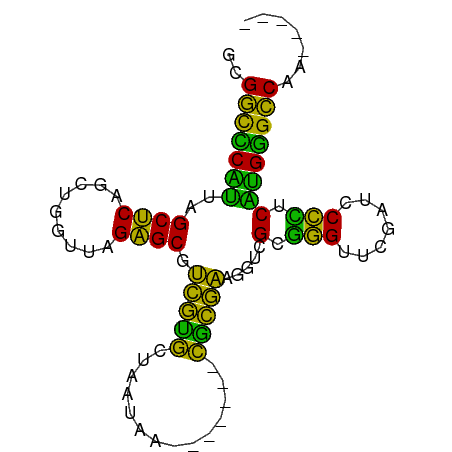
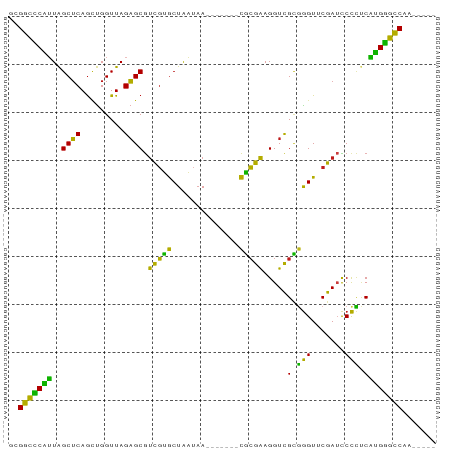
| Location | 9,325,368 – 9,325,445 |
|---|---|
| Length | 77 |
| Sequences | 14 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 83.82 |
| Shannon entropy | 0.36292 |
| G+C content | 0.58643 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -25.91 |
| Energy contribution | -24.74 |
| Covariance contribution | -1.17 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9325368 77 - 21146708 -----UUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCGC -----..(((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))).. ( -28.00, z-score = -2.35, R) >droGri2.scaffold_15112 1241212 81 + 5172618 -UGAUGUGGCCCAUGGGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCGA -.....((((((((.(((((.(((......))))))))((.-------......)).....((((.........))))..)))))))). ( -28.80, z-score = -0.97, R) >droMoj3.scaffold_6496 13663476 76 - 26866924 ------UGGCCCAUGGGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCAU ------((((((((.(((((.(((......))))))))((.-------......)).....((((.........))))..)))))))). ( -29.10, z-score = -1.81, R) >droVir3.scaffold_12875 1837644 82 - 20611582 UGCUGCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGG-------UUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCAC .......(((.((((((((.......))))).....(((((-------(.....)))))).((((.........))))..))).))).. ( -31.30, z-score = -2.14, R) >droWil1.scaffold_180772 6390000 77 - 8906247 -----AUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCUU -----..(((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))).. ( -27.50, z-score = -2.57, R) >dp4.chr3 6230985 78 + 19779522 ----UCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCAC ----..((((((((..(((.......)))(((.....))).-------.............((((.........))))..)))))))). ( -28.50, z-score = -2.91, R) >droPer1.super_2 8071538 78 - 9036312 ----UCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCAC ----..((((((((..(((.......)))(((.....))).-------.............((((.........))))..)))))))). ( -28.50, z-score = -2.91, R) >droAna3.scaffold_13266 16626120 77 - 19884421 -----CUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCGU -----..(((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).))))))))))).. ( -28.20, z-score = -1.72, R) >droEre2.scaffold_4845 6097252 77 + 22589142 -----UUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCGC -----..(((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).))))))))))).. ( -28.20, z-score = -1.80, R) >droYak2.chr2R 13505628 77 + 21139217 -----UUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCGC -----..(((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).))))))))))).. ( -28.20, z-score = -1.80, R) >droSec1.super_1 6844491 77 - 14215200 -----UUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCGC -----..(((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).))))))))))).. ( -28.20, z-score = -1.80, R) >droSim1.chr2R 7772970 77 - 19596830 -----UUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCGU -----..(((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).))))))))))).. ( -28.20, z-score = -1.80, R) >triCas2.chrUn_43 98017 83 + 275627 -----UUGGCUCAUGUAGGAUUCGAACCUACGACCUCCGCAAGGCUGCUGCUUAGUGCGGUGCUCUAACCACCUGAGCUAACGAACCG- -----((((((((.(((((.......)))))((.(.(((((.(((....)))...))))).).))........)))))))).......- ( -28.60, z-score = -2.73, R) >anoGam1.chr2R 13948018 74 + 62725911 ------UGGCUCCAGAGAGGAUCGAACUCACGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAUGGAGCC-- ------.(((((((..((((.(((......))).))))((.-------......)).....((((.........))))..)))))))-- ( -25.30, z-score = -2.26, R) >consensus _____UUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG_______UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCGC .......(((((((..(((.......)))(((.....))).....................((((.........))))..))))))).. (-25.91 = -24.74 + -1.17)

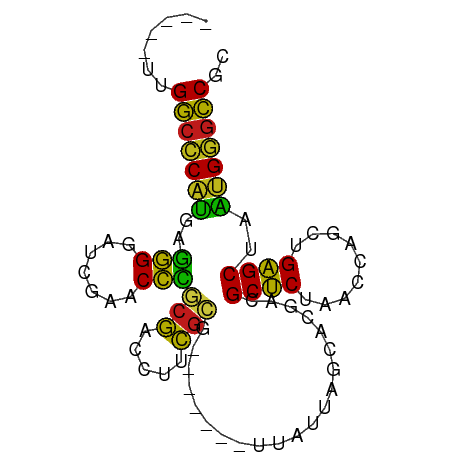
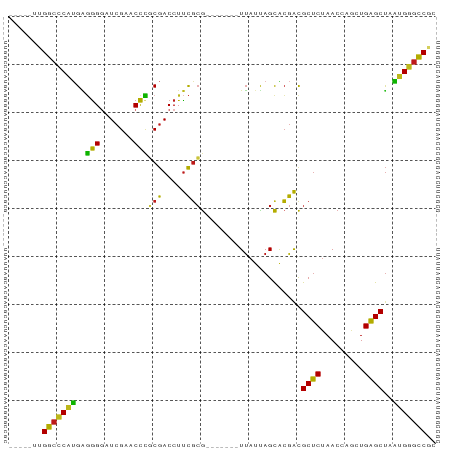
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:04 2011