
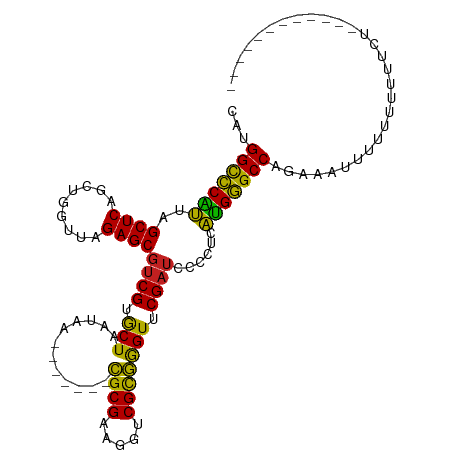
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,319,278 – 9,319,379 |
| Length | 101 |
| Max. P | 0.988250 |

| Location | 9,319,278 – 9,319,379 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.35 |
| Shannon entropy | 0.48122 |
| G+C content | 0.51434 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -26.81 |
| Energy contribution | -25.81 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9319278 101 + 21146708 UAUGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGAUAUUUUUUAUUUCUGAACAUCC---- ..((((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))))........................---- ( -29.20, z-score = -1.05, R) >droGri2.scaffold_15245 203951 92 + 18325388 CAUGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGCCGCGGGUUCGAUCCCCCCAUGGGCCAGGAUUGUCUUCUUUU------------- ..((((((((.......((.((((...((((..(....)..)-------)))...))))))(((......)))...))))))))(((......)))...------------- ( -30.90, z-score = -0.51, R) >droMoj3.scaffold_6496 25288206 90 + 26866924 UGUGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCCCAUGGGCCAAGAGU-UCUUAGUU-------------- ..((((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....)))))).....-........-------------- ( -29.50, z-score = -0.70, R) >droVir3.scaffold_12875 15829587 96 - 20611582 CGGGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGCCGCGGGUUCGAUCCCCCCAUGGGCCAAUUUAUAUUUUUCAAUUUU--------- ...(((((((.......((.((((...((((..(....)..)-------)))...))))))(((......)))...)))))))....................--------- ( -29.80, z-score = -0.35, R) >droPer1.super_2 8066107 92 + 9036312 UGUGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGAAACUACUUUUUUC------------- (.((((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....)))))).).............------------- ( -29.70, z-score = -1.03, R) >dp4.chr3 6225561 92 - 19779522 UGUGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGAAAAUACUUUUUUC------------- ..((((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....))))))(((((.....)))))------------- ( -30.30, z-score = -1.41, R) >droEre2.scaffold_4845 6091128 95 - 22589142 CAUGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGGAAACCUUUUUUUUUAA---------- ..((((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))))(....)............---------- ( -30.00, z-score = -1.23, R) >droYak2.chr2R 13499346 97 - 21139217 CAUGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGGAUUCUUUUUUAUUUUGAU-------- ..((((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....))))))....................-------- ( -29.60, z-score = -0.69, R) >droSec1.super_1 6837595 96 + 14215200 CAUGGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGACAAUUUUUAUUUUUGAA--------- ..((((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))))...................--------- ( -29.20, z-score = -1.27, R) >anoGam1.chr2R 13947286 102 - 62725911 ---GGCUCCAUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGUGAGUUCGAUCCUCUCUGGAGCCAUAGAGAAUGUUUUUAUAUUCCAAGCUAG ---(((((((((((...))))...((((.((((.(((....(-------(((....).))).))).)))).)))).))))))).(((.((((((....))))))....))). ( -34.30, z-score = -2.33, R) >droAna3.scaffold_13266 10206574 92 - 19884421 ---GGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAUA---AAUAUUUUGCUAUUAC------- ---(((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......)))))))...---................------- ( -28.20, z-score = -1.20, R) >triCas2.chrUn_43 98015 94 - 275627 GUCGGUUCGUUAGCUCAGGUGGUUAGAGCACCGCACUAAGCAGCAGCCUUGCGGAGGUCGUAGGUUCGAAUCCUACAUGAGCCAAACUUUUUUU------------------ ...((((((((.(((.(((((((......))))).)).)))))).(((((...))))).(((((.......)))))..)))))...........------------------ ( -29.80, z-score = -1.57, R) >consensus CAUGGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA_______CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCCAGAAAUUUUUUUUUUCU____________ ...(((((((..((((.........))))((((.(((............((((.....))))))).))))......)))))))............................. (-26.81 = -25.81 + -1.00)

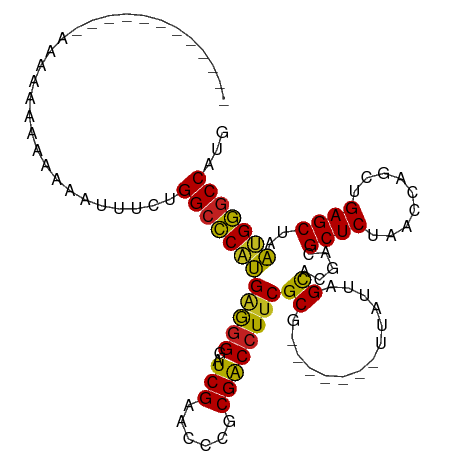
| Location | 9,319,278 – 9,319,379 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.35 |
| Shannon entropy | 0.48122 |
| G+C content | 0.51434 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -25.53 |
| Energy contribution | -24.99 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
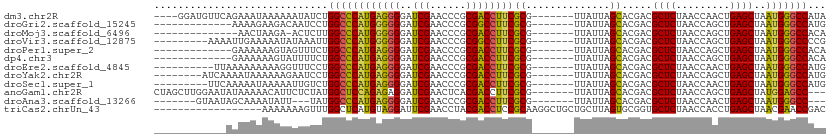
>dm3.chr2R 9319278 101 - 21146708 ----GGAUGUUCAGAAAUAAAAAAUAUCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCAUA ----(((((((...........)))))))(((((((..(((.......)))(((.....))).-------.............((((.........))))..)))))))... ( -30.70, z-score = -2.25, R) >droGri2.scaffold_15245 203951 92 - 18325388 -------------AAAAGAAGACAAUCCUGGCCCAUGGGGGGAUCGAACCCGCGGCCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCAUG -------------...............(((((((((.(((.......))).).....(((.(-------(.....)).))).((((.........))))..)))))))).. ( -28.70, z-score = -0.44, R) >droMoj3.scaffold_6496 25288206 90 - 26866924 --------------AACUAAGA-ACUCUUGGCCCAUGGGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCACA --------------........-.....((((((((.(((((.(((......))))))))((.-------......)).....((((.........))))..)))))))).. ( -29.20, z-score = -1.55, R) >droVir3.scaffold_12875 15829587 96 + 20611582 ---------AAAAUUGAAAAAUAUAAAUUGGCCCAUGGGGGGAUCGAACCCGCGGCCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCCCG ---------....................((((((((.(((.......))).).....(((.(-------(.....)).))).((((.........))))..)))))))... ( -27.20, z-score = -0.05, R) >droPer1.super_2 8066107 92 - 9036312 -------------GAAAAAAGUAGUUUCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCACA -------------...............((((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).)))))))))))).. ( -28.70, z-score = -1.39, R) >dp4.chr3 6225561 92 + 19779522 -------------GAAAAAAGUAUUUUCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCACA -------------(((((.....)))))((((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).)))))))))))).. ( -29.70, z-score = -2.09, R) >droEre2.scaffold_4845 6091128 95 + 22589142 ----------UUAAAAAAAAAGGUUUCCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCAUG ----------..................((((((((..(((.......)))(((.....))).-------.............((((.........))))..)))))))).. ( -28.60, z-score = -1.93, R) >droYak2.chr2R 13499346 97 + 21139217 --------AUCAAAAUAAAAAAGAAUCCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCAUG --------....................((((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).)))))))))))).. ( -28.80, z-score = -2.09, R) >droSec1.super_1 6837595 96 - 14215200 ---------UUCAAAAAUAAAAAUUGUCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCCAUG ---------...................((((((((..(((.......)))(((.....))).-------.............((((.........))))..)))))))).. ( -28.60, z-score = -2.44, R) >anoGam1.chr2R 13947286 102 + 62725911 CUAGCUUGGAAUAUAAAAACAUUCUCUAUGGCUCCAGAGAGGAUCGAACUCACGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAUGGAGCC--- .......................(((((((((((.((.((((.(((......))).))))(((-------((........))))).........)))))))))))))..--- ( -29.20, z-score = -2.03, R) >droAna3.scaffold_13266 10206574 92 + 19884421 -------GUAAUAGCAAAAUAUU---UAUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCC--- -------................---...(((((((..(((.......)))(((.....))).-------.............((((.........))))..)))))))--- ( -26.30, z-score = -1.54, R) >triCas2.chrUn_43 98015 94 + 275627 ------------------AAAAAAAGUUUGGCUCAUGUAGGAUUCGAACCUACGACCUCCGCAAGGCUGCUGCUUAGUGCGGUGCUCUAACCACCUGAGCUAACGAACCGAC ------------------.......(((.((((((.(((((.......)))))((.(.(((((.(((....)))...))))).).))........)))))))))........ ( -28.90, z-score = -2.37, R) >consensus ____________AAAAAAAAAAAAUUUCUGGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG_______UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCCAUG .............................(((((((..(((.......)))(((.....))).....................((((.........))))..)))))))... (-25.53 = -24.99 + -0.54)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:03 2011