
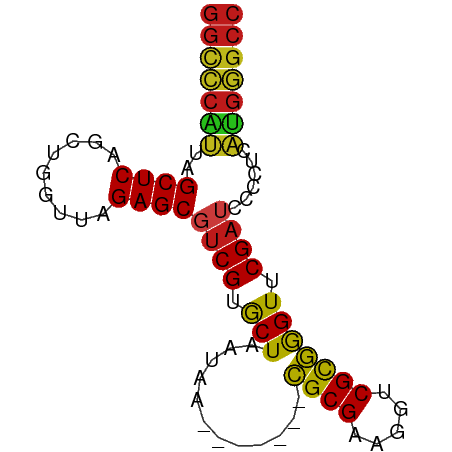
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,318,789 – 9,318,862 |
| Length | 73 |
| Max. P | 0.959121 |

| Location | 9,318,789 – 9,318,862 |
|---|---|
| Length | 73 |
| Sequences | 14 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 89.99 |
| Shannon entropy | 0.23672 |
| G+C content | 0.59015 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -23.24 |
| Energy contribution | -22.99 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
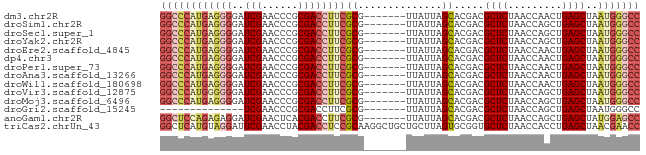
>dm3.chr2R 9318789 73 + 21146708 GGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCC (((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))) ( -26.50, z-score = -1.37, R) >droSim1.chr2R 7772393 73 + 19596830 GGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCC (((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....))))) ( -26.90, z-score = -1.13, R) >droSec1.super_1 6837149 73 + 14215200 GGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCC (((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....))))) ( -26.90, z-score = -1.13, R) >droYak2.chr2R 13498585 73 - 21139217 GGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCC (((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....))))) ( -26.90, z-score = -1.13, R) >droEre2.scaffold_4845 6090353 73 - 22589142 GGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCC (((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))) ( -26.50, z-score = -1.37, R) >dp4.chr3 1803075 73 - 19779522 GGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCC (((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))) ( -26.50, z-score = -1.37, R) >droPer1.super_73 80096 73 + 267513 GGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCC (((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))) ( -26.50, z-score = -1.37, R) >droAna3.scaffold_13266 10206413 73 - 19884421 GGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCC (((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))) ( -26.50, z-score = -1.37, R) >droWil1.scaffold_180698 1923958 73 + 11422946 GGCCCAUUAGCUCAGUUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCC (((((((..((((.........))))((((.(((.....-------((((.....))))))).))))......))))))) ( -26.50, z-score = -1.37, R) >droVir3.scaffold_12875 15829282 73 - 20611582 GGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCCCAUGGGCC (((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....))))) ( -26.90, z-score = -0.94, R) >droMoj3.scaffold_6496 13658315 73 + 26866924 GGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCC (((((((((((.(.(((......)))...).))))))..-------.(((.....)))(((......))).....))))) ( -26.90, z-score = -1.13, R) >droGri2.scaffold_15245 203679 59 + 18325388 GGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGCGGGUUCGA-------------- (((((((((((.(.(((......)))...).))))))..-------.(((.....))))))))...-------------- ( -19.70, z-score = -0.61, R) >anoGam1.chr2R 13946496 73 - 62725911 GGCUCCAUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA-------CGCGAAGGUCGUGAGUUCGAUCCUCUCUGGAGCC (((((((((((...))))...((((.((((.(((....(-------(((....).))).))).)))).)))).))))))) ( -28.80, z-score = -2.38, R) >triCas2.chrUn_43 98018 80 - 275627 GGUUCGUUAGCUCAGGUGGUUAGAGCACCGCACUAAGCAGCAGCCUUGCGGAGGUCGUAGGUUCGAAUCCUACAUGAGCC ((((((((.(((.(((((((......))))).)).)))))).(((((...))))).(((((.......)))))..))))) ( -27.80, z-score = -1.53, R) >consensus GGCCCAUUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAA_______CGCGAAGGUCGCGGGUUCGAUCCCCUCAUGGGCC (((((((..((((.........))))((((.(((............((((.....))))))).))))......))))))) (-23.24 = -22.99 + -0.26)

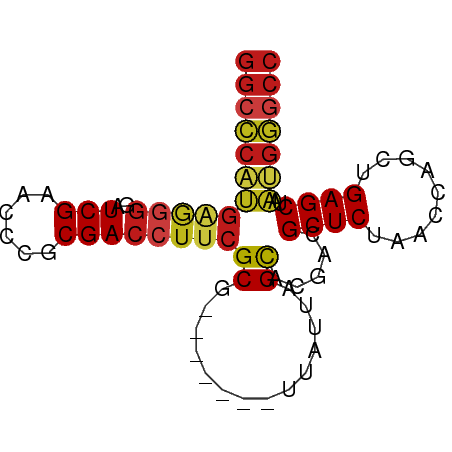
| Location | 9,318,789 – 9,318,862 |
|---|---|
| Length | 73 |
| Sequences | 14 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 89.99 |
| Shannon entropy | 0.23672 |
| G+C content | 0.59015 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -21.79 |
| Energy contribution | -22.09 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9318789 73 - 21146708 GGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCC (((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))) ( -26.30, z-score = -2.45, R) >droSim1.chr2R 7772393 73 - 19596830 GGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCC (((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).))))))))))) ( -26.50, z-score = -1.92, R) >droSec1.super_1 6837149 73 - 14215200 GGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCC (((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).))))))))))) ( -26.50, z-score = -1.92, R) >droYak2.chr2R 13498585 73 + 21139217 GGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCC (((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).))))))))))) ( -26.50, z-score = -1.92, R) >droEre2.scaffold_4845 6090353 73 + 22589142 GGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCC (((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))) ( -26.30, z-score = -2.45, R) >dp4.chr3 1803075 73 + 19779522 GGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCC (((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))) ( -26.30, z-score = -2.45, R) >droPer1.super_73 80096 73 - 267513 GGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCC (((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))) ( -26.30, z-score = -2.45, R) >droAna3.scaffold_13266 10206413 73 + 19884421 GGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCC (((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))) ( -26.30, z-score = -2.45, R) >droWil1.scaffold_180698 1923958 73 - 11422946 GGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAACUGAGCUAAUGGGCC (((((((..(((.......)))(((.....))).-------.............((((.........))))..))))))) ( -26.30, z-score = -2.45, R) >droVir3.scaffold_12875 15829282 73 + 20611582 GGCCCAUGGGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCC (((((((.(((((.(((......))))))))((.-------......)).....((((.........))))..))))))) ( -26.90, z-score = -1.36, R) >droMoj3.scaffold_6496 13658315 73 - 26866924 GGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCC (((((((..(((.......)))(((.....))).-------....((((.((..(((......))))).))))))))))) ( -26.50, z-score = -1.92, R) >droGri2.scaffold_15245 203679 59 - 18325388 --------------UCGAACCCGCGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCC --------------.....((((((.....))).-------..((((((.((..(((......))))).))))))))).. ( -16.00, z-score = -1.10, R) >anoGam1.chr2R 13946496 73 + 62725911 GGCUCCAGAGAGGAUCGAACUCACGACCUUCGCG-------UUAUUAGCACGACGCUCUAACCAGCUGAGCUAUGGAGCC (((((((..((((.(((......))).))))((.-------......)).....((((.........))))..))))))) ( -25.30, z-score = -2.40, R) >triCas2.chrUn_43 98018 80 + 275627 GGCUCAUGUAGGAUUCGAACCUACGACCUCCGCAAGGCUGCUGCUUAGUGCGGUGCUCUAACCACCUGAGCUAACGAACC ((((((.(((((.......)))))((.(.(((((.(((....)))...))))).).))........))))))........ ( -26.90, z-score = -2.37, R) >consensus GGCCCAUGAGGGGAUCGAACCCGCGACCUUCGCG_______UUAUUAGCACGACGCUCUAACCAGCUGAGCUAAUGGGCC (((((((..(((.......)))(((.....))).....................((((.........))))..))))))) (-21.79 = -22.09 + 0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:20:01 2011