| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,315,192 – 9,315,305 |
| Length | 113 |
| Max. P | 0.998772 |

| Location | 9,315,192 – 9,315,305 |
|---|---|
| Length | 113 |
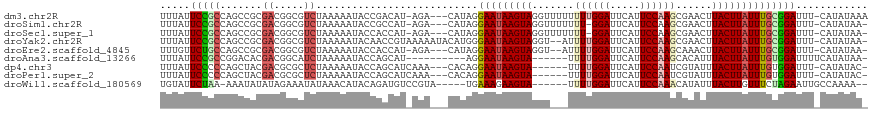
| Sequences | 9 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.01 |
| Shannon entropy | 0.41769 |
| G+C content | 0.35890 |
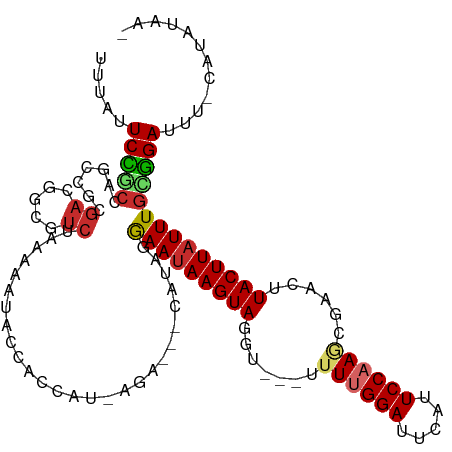
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -14.67 |
| Energy contribution | -15.14 |
| Covariance contribution | 0.47 |
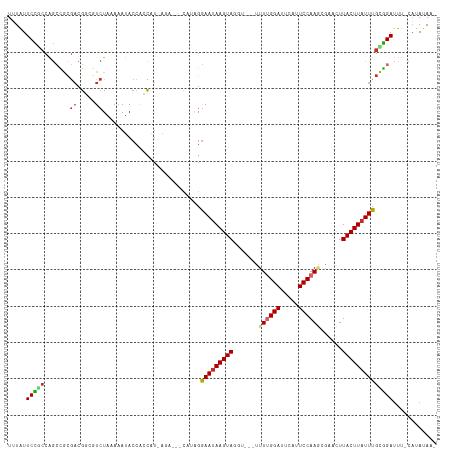
| Combinations/Pair | 1.27 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.998772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9315192 113 + 21146708 UUUAUUCCGCCAGCCGCGACGGCGUCUAAAAAUACCGACAU-AGA---CAUAGGAAUAAGUAGGUUUUUUUUGGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUUU-CAUAUAAA .....(((((..((((...))))(((((............)-)))---)....((((((((((((((..((((((.....)))))).)))))))))))))))))))...-........ ( -36.40, z-score = -5.15, R) >droSim1.chr2R 7768824 111 + 19596830 UUUAUUCCGCCAGCCGCGACGGCGUCUAAAAAUACCGCCAU-AGA---CAUAGGAAUAAGUAGGUUUUUUU-GGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUUU-CAUAUAA- .....(((((..((((...))))(((((............)-)))---)....((((((((((((((.(((-(((.....)))))).)))))))))))))))))))...-.......- ( -38.40, z-score = -5.48, R) >droSec1.super_1 6833674 111 + 14215200 UUUAUUCCGCCAGCCGCGACGGCGUCUAAAAAUACCACCAU-AGA---CAUAGGAAUAAGUAGGUUUUUUU-GGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUUU-CAUAUAA- .....(((((..((((...))))(((((............)-)))---)....((((((((((((((.(((-(((.....)))))).)))))))))))))))))))...-.......- ( -38.40, z-score = -6.21, R) >droYak2.chr2R 13494063 114 - 21139217 UUUAUUCCGCCAGCCGCGACGGCGUCUAAAAAUACAACCGUAAAAAUACAUGGGAAUAAGUAGGU--AUUUUGGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUUU-CAUAUAA- .....(((((..((((...))))..............((((........))))((((((((((((--..((((((.....))))))...)))))))))))))))))...-.......- ( -33.20, z-score = -3.66, R) >droEre2.scaffold_4845 6086536 110 - 22589142 UUUGUUCUGCCAGCCGCGACGGCGUCUAAAAAUACCACCAU-AGA---CAUAGGAAUAAGUAGGU--AUUUUGGAUUCAUUCCAAGCAAACUUACUUAUUUGCGGAUUU-CAUAUAA- ..((.(((((..((((...))))(((((............)-)))---)....((((((((((((--..((((((.....))))))...)))))))))))))))))...-)).....- ( -32.10, z-score = -3.96, R) >droAna3.scaffold_13266 16617517 101 + 19884421 UUUAUUCCGCCGGACACGACGGCAUCUAAAAAUACCAGCAU----------AGGAAUAAGUA------UUUUGGAUUCAUUCCAAGCACAUUUACUUAUUUGUGGAUUUUCAUAUAA- .....(((((((.......))))..................----------(.(((((((((------.((((((.....))))))......))))))))).))))...........- ( -22.00, z-score = -2.66, R) >dp4.chr3 6222232 107 - 19779522 UUUAUUCCCCCAGCUACGACGCGCUCUAAAAAUACCAGCAUCAAA---CACAGGAAUAAGUA------UUUUGGAUUCAUUCCAAUCGUAUUUACUUAUUUGUGGAUUU-CAUAUAC- ............((......))(((...........)))......---..((.(((((((((------..(((((.....))))).......))))))))).)).....-.......- ( -17.00, z-score = -1.30, R) >droPer1.super_2 8062775 107 + 9036312 UUUAUUCCCCCAGCUACGACGCGCUCUAAAAAUACCAGCAUCAAA---CACAGGAAUAAGUA------UUUUGGAUUCAUUCCAAUCGUAUUUACUUAUUUGUGGAUUU-CAUAUAC- ............((......))(((...........)))......---..((.(((((((((------..(((((.....))))).......))))))))).)).....-.......- ( -17.00, z-score = -1.30, R) >droWil1.scaffold_180569 469048 104 - 1405142 UGUAUUCUAA-AAAUAUAUAGAAAUAUAAACAUACAGAUGUCCGUA-----UGAAAGAAGUA------UUUUGGAUUCAUUCCAAACAUAUUUACUUGUUUCUAGAAUUGCCAAAA-- .((((((((.-((((.((((....))))..(((((.(....).)))-----)).((((((((------(((((((.....)))))).)))))).))))))).))))).))).....-- ( -21.00, z-score = -2.98, R) >consensus UUUAUUCCGCCAGCCGCGACGGCGUCUAAAAAUACCACCAU_AGA___CAUAGGAAUAAGUAGGU___UUUUGGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUUU_CAUAUAA_ .....(((((.......((.....))...........................(((((((((.......((((((.....))))))......))))))))))))))............ (-14.67 = -15.14 + 0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:58 2011