| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,252,630 – 9,252,737 |
| Length | 107 |
| Max. P | 0.517720 |

| Location | 9,252,630 – 9,252,737 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.18 |
| Shannon entropy | 0.39984 |
| G+C content | 0.49502 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -15.36 |
| Energy contribution | -16.23 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
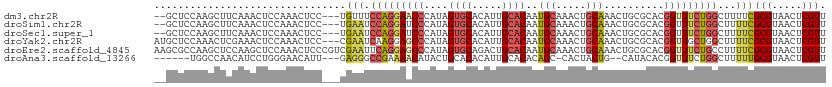
>dm3.chr2R 9252630 107 - 21146708 --GCUCCAAGCUUCAAACUCCAAACUCC---UGUUUCCAGGAACCCAUAGUGCACAUUGCACAAUGCAAACUGCAAACUGCGCACGGUUUCUGGCUUUUCGCGUAACUCGUU --((...(((((..(((((......(((---((....))))).......((((...((((.....))))...((.....)))))))))))..)))))...)).......... ( -27.60, z-score = -2.22, R) >droSim1.chr2R 7710093 107 - 19596830 --GCUCCAAGCUUCAAACUCCAAACUCC---UGAAUCCAGGAUCCCAUAGUGCACAUUGCACAAUGCAAACUGCAAACUGCGCACGGUUUCUGGCUUUUCGCGUAACUCGUU --((...(((((..(((((......(((---((....))))).......((((...((((.....))))...((.....)))))))))))..)))))...)).......... ( -27.90, z-score = -2.45, R) >droSec1.super_1 6775629 107 - 14215200 --GCUCCAAGCUUCAAACUCCAAACUCC---UGAAUCCAGGAUCCCAUAGUGCACAUUGCACAAUGCAAACUGCAAACUGCGCACGGUUUCUGGCUUUUCGCGUAACUCGUU --((...(((((..(((((......(((---((....))))).......((((...((((.....))))...((.....)))))))))))..)))))...)).......... ( -27.90, z-score = -2.45, R) >droYak2.chr2R 13434545 109 + 21139217 AUGCUCCAAACUCGAAACUCCAAACUCC---CGAAUCAAGGAGCCCAUAGUGCACAUUGCACAAUGCAAACUGCAAACUGCGCACGGUUGCUGGCUUUUCGCGUAACUCGUU ..(((((....(((..............---))).....))))).....(((((..(((((..........)))))..)))))((((((((..((.....))))))).))). ( -25.24, z-score = -0.41, R) >droEre2.scaffold_4845 6026314 112 + 22589142 AAGCGCCAAGCUCCAAGCUCCAAACUCCCGUCGAAUUCAGGAGCCCAUAGUGCAGACUGCACAAUGCAAACUGCAAACUGCGCACGGUUUCUGCCUUUUCGCGUAACUCGUU ..((((..(((.....))).............(((..((((((((....((((((..((((..........))))..))))))..))))))))....)))))))........ ( -30.30, z-score = -1.03, R) >droAna3.scaffold_13266 16560381 100 - 19884421 ------UGGCCAACAUCCUGGGAACAUU---GAGGGCCGAAAACAUACUGCACACAUUGCACACAGC-CACUACUG--CAUACACGGUUUCUGGCUUUUUGCGUAACUCGUU ------.((((..((...((....)).)---)..))))....................(((.(.(((-((..((((--......))))...))))).).))).......... ( -23.90, z-score = -0.50, R) >consensus __GCUCCAAGCUUCAAACUCCAAACUCC___UGAAUCCAGGAACCCAUAGUGCACAUUGCACAAUGCAAACUGCAAACUGCGCACGGUUUCUGGCUUUUCGCGUAACUCGUU ................................(((.(((((((((....((((.....))))..(((.....)))..........)))))))))...)))(((.....))). (-15.36 = -16.23 + 0.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:52 2011