| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,249,337 – 9,249,438 |
| Length | 101 |
| Max. P | 0.944867 |

| Location | 9,249,337 – 9,249,438 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 86.19 |
| Shannon entropy | 0.27403 |
| G+C content | 0.37473 |
| Mean single sequence MFE | -24.99 |
| Consensus MFE | -21.79 |
| Energy contribution | -21.93 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821716 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
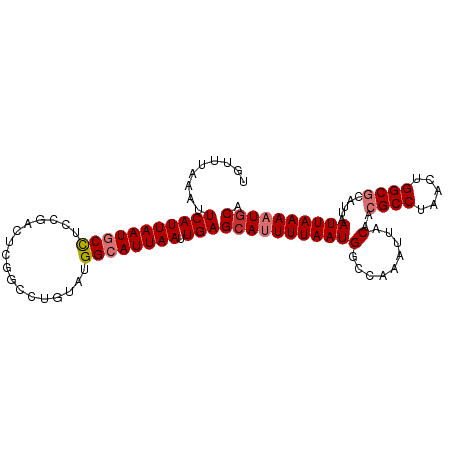
>dm3.chr2R 9249337 101 + 21146708 UGUAUUUUAAUUAAUGCGCCAGUUAGGCGUUGUAAUUUCGCCAUUAAAAUGCUCAAUUAAUGCCAUACAGGCCGAGUCGGAGGCAUUAAUGAAUUUAAACA .((((((((((((((......))))((((.........))))))))))))))(((.((((((((.....(((...)))...)))))))))))......... ( -24.00, z-score = -1.17, R) >droSim1.chr2R 7706843 101 + 19596830 UGCAUUUUAAUUAAUGCGCCAGUUAGGCGUUGUAAUUUGGCCAUUAAAAUGCUCAAUUAAUGCCAUACAGGCCGAGUCGCAGGCAUUAAUGAAUUUAAACA .........(((((((((((.....))).(((..((((((((........((.........))......))))))))..)))))))))))........... ( -27.54, z-score = -1.70, R) >droSec1.super_1 6772383 101 + 14215200 UGCAUUUUAAUUAAUGCGCCAGUUAGGCGUUGUAAUUUGGCCAUUAAAAUGCUCAAUUAAUGCCAUACAGGCCGAGUCGCAGGCAUUAAUGAAUUUAAACA .........(((((((((((.....))).(((..((((((((........((.........))......))))))))..)))))))))))........... ( -27.54, z-score = -1.70, R) >droYak2.chr2R 13430755 101 - 21139217 UGCAUUUUAAUUAAUGCGCCAGUUAGGCGUUGUAAUUUGGCCAUUAAAAUGCUCAAUUAAUGCCAUGCAGGCCGAGUCGGAGGCAUUAAUGAAUUUAAACG .((((((((((....(((((.....))))).((......)).))))))))))(((.((((((((...(.(((...))).).)))))))))))......... ( -26.40, z-score = -0.88, R) >droEre2.scaffold_4845 6022919 101 - 22589142 UGCAUUUUAAUUAAUGCGCCAGUUAGGCGUUGUAAUUUGGCCAUUAAAAUGCUCAAUUAAUGCCAUACAGGGCGAGUCGGAGGCAUUAAUGAAUUUAAACA .((((((((((....(((((.....))))).((......)).))))))))))(((.((((((((.................)))))))))))......... ( -24.53, z-score = -0.70, R) >droAna3.scaffold_13266 16557117 86 + 19884421 AGCAUUUUAAUUAAUGCGCCAGUUAGGCGUUGUAAUUCUGCCAUUAAAAUGCUCAAUUAAUGCCA---------------GAGCAUUAAUGAAUUUACACG .((((((((((....(((((.....))))).(((....))).))))))))))(((.(((((((..---------------..))))))))))......... ( -23.10, z-score = -2.94, R) >dp4.chr3 19118038 95 - 19779522 CGCAGUUUAAUUAAUGUGCCAGUUAGGCGAUGUAAUUUUGCCAUUAAAAUGCUCAUUUAAUGCCGAGCGAG------CGGAGGUAUUAAUGAAUUUUAGCA .((.((..(((((...((((.....))))...)))))..))...((((((..(((((.((((((..(....------)...))))))))))))))))))). ( -23.40, z-score = -0.93, R) >droPer1.super_2 8208063 95 - 9036312 CGCAGUUUAAUUAAUGUGCCAGUUAGGCGAUGUAAUUUUGCCAUUAAAAUGCUCAUUUAAUGCCGAGCGAG------CGGAGGUAUUAAUGAAUUUUAGCA .((.((..(((((...((((.....))))...)))))..))...((((((..(((((.((((((..(....------)...))))))))))))))))))). ( -23.40, z-score = -0.93, R) >consensus UGCAUUUUAAUUAAUGCGCCAGUUAGGCGUUGUAAUUUGGCCAUUAAAAUGCUCAAUUAAUGCCAUACAGGCCGAGUCGGAGGCAUUAAUGAAUUUAAACA .((((((((((....(((((.....))))).((......)).))))))))))(((.((((((((.................)))))))))))......... (-21.79 = -21.93 + 0.14)
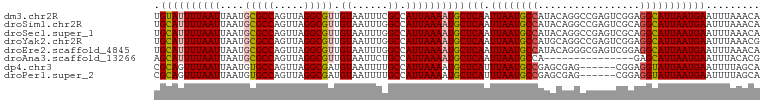
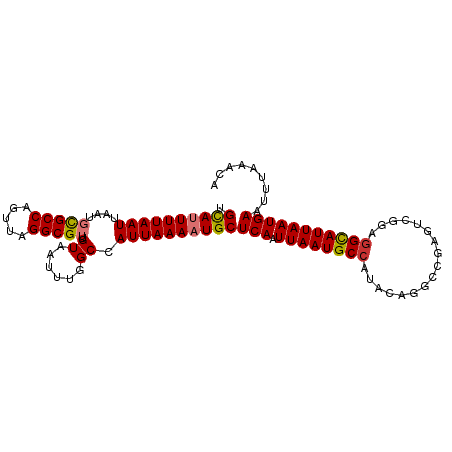
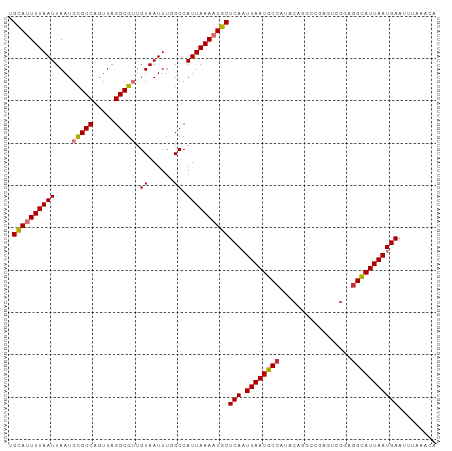
| Location | 9,249,337 – 9,249,438 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 86.19 |
| Shannon entropy | 0.27403 |
| G+C content | 0.37473 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -19.84 |
| Energy contribution | -20.61 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944867 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 9249337 101 - 21146708 UGUUUAAAUUCAUUAAUGCCUCCGACUCGGCCUGUAUGGCAUUAAUUGAGCAUUUUAAUGGCGAAAUUACAACGCCUAACUGGCGCAUUAAUUAAAAUACA .(((((.....(((((((((....((.......))..))))))))))))))((((((((.(((......)...(((.....)))))....))))))))... ( -22.00, z-score = -1.31, R) >droSim1.chr2R 7706843 101 - 19596830 UGUUUAAAUUCAUUAAUGCCUGCGACUCGGCCUGUAUGGCAUUAAUUGAGCAUUUUAAUGGCCAAAUUACAACGCCUAACUGGCGCAUUAAUUAAAAUGCA .........(((((((((((((((........)))).)))))))).)))(((((((((((.........)..((((.....)))).....)))))))))). ( -27.90, z-score = -2.35, R) >droSec1.super_1 6772383 101 - 14215200 UGUUUAAAUUCAUUAAUGCCUGCGACUCGGCCUGUAUGGCAUUAAUUGAGCAUUUUAAUGGCCAAAUUACAACGCCUAACUGGCGCAUUAAUUAAAAUGCA .........(((((((((((((((........)))).)))))))).)))(((((((((((.........)..((((.....)))).....)))))))))). ( -27.90, z-score = -2.35, R) >droYak2.chr2R 13430755 101 + 21139217 CGUUUAAAUUCAUUAAUGCCUCCGACUCGGCCUGCAUGGCAUUAAUUGAGCAUUUUAAUGGCCAAAUUACAACGCCUAACUGGCGCAUUAAUUAAAAUGCA .........(((((((((((.(((...))).......)))))))).)))(((((((((((.........)..((((.....)))).....)))))))))). ( -25.20, z-score = -1.74, R) >droEre2.scaffold_4845 6022919 101 + 22589142 UGUUUAAAUUCAUUAAUGCCUCCGACUCGCCCUGUAUGGCAUUAAUUGAGCAUUUUAAUGGCCAAAUUACAACGCCUAACUGGCGCAUUAAUUAAAAUGCA .........(((((((((((....((.......))..)))))))).)))(((((((((((.........)..((((.....)))).....)))))))))). ( -26.20, z-score = -2.76, R) >droAna3.scaffold_13266 16557117 86 - 19884421 CGUGUAAAUUCAUUAAUGCUC---------------UGGCAUUAAUUGAGCAUUUUAAUGGCAGAAUUACAACGCCUAACUGGCGCAUUAAUUAAAAUGCU ...(((..((.((((((((((---------------((.((((((.........)))))).))))........(((.....))))))))))).))..))). ( -24.00, z-score = -2.95, R) >dp4.chr3 19118038 95 + 19779522 UGCUAAAAUUCAUUAAUACCUCCG------CUCGCUCGGCAUUAAAUGAGCAUUUUAAUGGCAAAAUUACAUCGCCUAACUGGCACAUUAAUUAAACUGCG ......................((------(..(((((........)))))...(((((((........)...(((.....))).)))))).......))) ( -15.40, z-score = -0.63, R) >droPer1.super_2 8208063 95 + 9036312 UGCUAAAAUUCAUUAAUACCUCCG------CUCGCUCGGCAUUAAAUGAGCAUUUUAAUGGCAAAAUUACAUCGCCUAACUGGCACAUUAAUUAAACUGCG ......................((------(..(((((........)))))...(((((((........)...(((.....))).)))))).......))) ( -15.40, z-score = -0.63, R) >consensus UGUUUAAAUUCAUUAAUGCCUCCGACUCGGCCUGUAUGGCAUUAAUUGAGCAUUUUAAUGGCCAAAUUACAACGCCUAACUGGCGCAUUAAUUAAAAUGCA .........(((((((((((.................)))))))).)))(((((((((((.........)..((((.....)))).....)))))))))). (-19.84 = -20.61 + 0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:51 2011