| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,247,096 – 9,247,189 |
| Length | 93 |
| Max. P | 0.937981 |

| Location | 9,247,096 – 9,247,189 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.04 |
| Shannon entropy | 0.54133 |
| G+C content | 0.60256 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.937981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9247096 93 + 21146708 -----CUCCCGCUGUGUGUGCGGUUUUGUUUGACUGCCACCAGGUGGCACCUGCCCC--------ACUAUCCACUAUCCGCAAGCCACUCCCUCUU--CCAGCACACC---------- -----.....((((.(((.((((((......)))))))))..((((((...(((...--------..............))).)))))).......--.)))).....---------- ( -25.23, z-score = -1.52, R) >droSim1.chr2R 7704557 91 + 19596830 -----CUCCCGCUGUGUGUGCGGUUUUGUUUGACUGCCACCAGGUGGCACCUGCCCC--------ACUAUCCGCUAUCCGCUAGCCACUCCCUCUU--CCAGCACC------------ -----.....((((.(((.((((((......)))))))))..((.(((....)))))--------.......((((.....))))...........--.))))...------------ ( -26.10, z-score = -1.87, R) >droSec1.super_1 6770082 91 + 14215200 -----CUCCCGCUGUGUGUGCGGUUUUGUUUGACUGCCACCAGGUGGCACCUGCCCC--------ACUAUCCGCUAUCCGCUAGCCACUCCCUCUU--CCAGCACC------------ -----.....((((.(((.((((((......)))))))))..((.(((....)))))--------.......((((.....))))...........--.))))...------------ ( -26.10, z-score = -1.87, R) >droYak2.chr2R 13428401 113 - 21139217 -----CUCCCGCUGUGUGUGCGGUUUUGUUUGACUGCCACCAGGUGGCACCUGCCCCCCUACCCUACAACCCACUAUCCAGAAUCCAGUAUCCACAAGCCACUCCCUCUUGCAGCACC -----.....((((((((.((((((......)))))))))..((((((..(((.........................)))......((....))..)))))).......)))))... ( -26.21, z-score = -0.95, R) >droEre2.scaffold_4845 6020611 104 - 22589142 -----CUCACGCUGUGUGCGCGGUUUUGUUUGACUGCCACCAGGUGGCACCUGCCCC--------ACCAUGCACCAUCCACCAUCCACUAUCCAGAAGCCACUGCCUCUC-CAGCACC -----.....((((.(((.((((((......)))))))))..(((((....(((...--------.....)))....)))))...........(((.((....)).))).-))))... ( -28.30, z-score = -1.04, R) >droAna3.scaffold_13266 16554521 81 + 19884421 CUCUGCACGCAGUUCUGGUGCGGUUUUGUUUGACUGCCACCAGGUGGCA--UGCCAC--------ACUCCCUACCUCCCCACUGGAGCUUC--------------------------- ..(((....)))..(((((((((((......))))).))))))((((..--..))))--------.........((((.....))))....--------------------------- ( -26.70, z-score = -1.24, R) >consensus _____CUCCCGCUGUGUGUGCGGUUUUGUUUGACUGCCACCAGGUGGCACCUGCCCC________ACUAUCCACUAUCCGCAAGCCACUCCCUCUU__CCAGCACC____________ ...........(((.(((.((((((......))))))))))))..(((....)))............................................................... (-15.68 = -16.35 + 0.67)

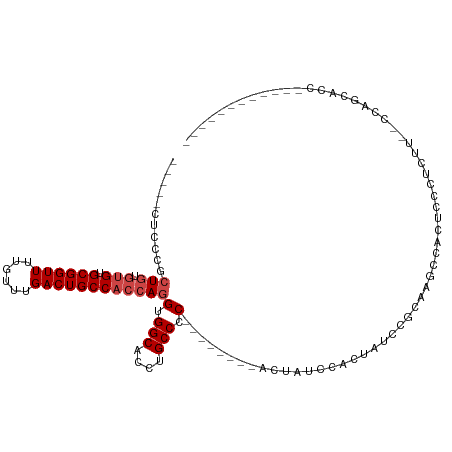
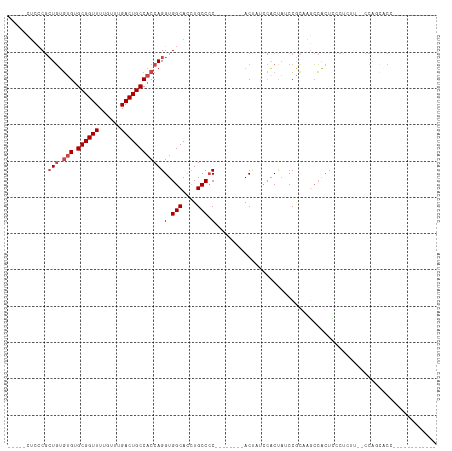
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:50 2011