| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,220,592 – 9,220,699 |
| Length | 107 |
| Max. P | 0.771005 |

| Location | 9,220,592 – 9,220,699 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.59 |
| Shannon entropy | 0.32278 |
| G+C content | 0.34179 |
| Mean single sequence MFE | -15.99 |
| Consensus MFE | -11.32 |
| Energy contribution | -11.26 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
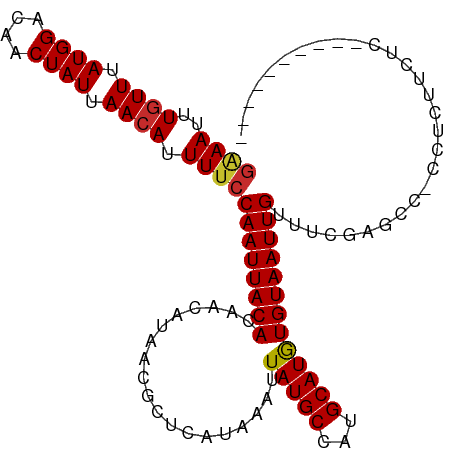
>dm3.chr2R 9220592 107 + 21146708 GAAAUUUGUUUAUGGACAACUAUUAACAUUUUCCAAUUACACAACAUAACGCUCAUAAAUUAUGCCAUGCAUGUGUAAUUGUUUCGAGCC-CCUUUUCCCUCCUCUCA-- ((((..((((.((((....)))).)))).))))((((((((((.(((...((...........)).)))..))))))))))....(((..-.............))).-- ( -16.96, z-score = -2.31, R) >droAna3.scaffold_13266 16528482 103 + 19884421 GAAAUUUGUUUAUGGACAACUAUUAACAUUUUCCAAUUACACAA---AACGCUCAUAAAUUAUGCCAUGCAUAUGUAAUUGCCUCG-CCC-CCUCUAAUCCAAUCGAA-- ((((..((((.((((....)))).)))).))))((((((((...---.............(((((...))))))))))))).....-...-.................-- ( -12.49, z-score = -1.03, R) >droEre2.scaffold_4845 5994553 97 - 22589142 GAAAUUUGUUUAUGGACAACUAUUAACAUUUUCCAAUUACACAACAUAACGCUCAUAAAUUAUGCCAUGCAUGUGUAAUUGUUUCGAGCC-CCGCUCA------------ ((((..((((.((((....)))).)))).))))((((((((((.(((...((...........)).)))..))))))))))....((((.-..)))).------------ ( -21.10, z-score = -2.97, R) >droYak2.chr2R 13400535 97 - 21139217 GAAAUUUGUUUAUGGACAACUAUUAACAUUUUCCAAUUACACAACAUAACGCUCAUAAAUUAUGCCAUGCAUGUGUAAUUGUUUCGAUCC-CCUCUUA------------ ((((..((((.((((....)))).)))).))))((((((((((.(((...((...........)).)))..)))))))))).........-.......------------ ( -16.90, z-score = -2.40, R) >droSec1.super_1 6743655 101 + 14215200 GAAAUUUGUUUAUGGACAACUAUUAACAUUUUCCAAUUACACAACAUAACGCUCAUAAAUUAUGCCAUGCAUGUGUAAUUGUUUCGAUCC-CCUCUUCCCUC-------- ((((..((((.((((....)))).)))).))))((((((((((.(((...((...........)).)))..)))))))))).........-...........-------- ( -16.90, z-score = -2.66, R) >droSim1.chr2R 7677981 101 + 19596830 GAAAUUUGUUUAUGGACAACUAUUAACAUUUUCCAAUUACACAACAUAACGCUCAUAAAUUAUGCCAUGCAUGUGUAAUUGUUUCGAUCC-CCUCUUCCCUC-------- ((((..((((.((((....)))).)))).))))((((((((((.(((...((...........)).)))..)))))))))).........-...........-------- ( -16.90, z-score = -2.66, R) >droPer1.super_2 8176955 96 - 9036312 GGAAUUUGUUUAUGGACAGCUAUUAACAUUUUCCAAUUACACAA---AACGCUCAUAAAUUAUGCCAUGCAUAUGUAAUUGUUUGUCGUC-CAGUCUCUC---------- ((((..((((.((((....)))).))))..))))(((((((...---.............(((((...))))))))))))..........-.........---------- ( -15.79, z-score = -0.72, R) >dp4.chr3 19086580 96 - 19779522 GGAAUUUGUUUAUGGACAGCUAUUAACAUUUUCCAAUUACACAA---AACGCUCAUAAAUUAUGCCAUGCAUAUGUAAUUGUUUGUCGUC-CAGUCUCUC---------- ((((..((((.((((....)))).))))..))))(((((((...---.............(((((...))))))))))))..........-.........---------- ( -15.79, z-score = -0.72, R) >droWil1.scaffold_181009 2077101 106 + 3585778 AAAAUUUGUUUAUGGACAACUAUUAAAAUUU-CCAAUUACACAA---AACGCUCAUAAAUUAUGCUAUGCAUAUGUAAUUGUUGCAAUCUGCCCCCAGUUUCAUUUCCCC ......((....(((................-.((((((((...---.............(((((...)))))))))))))..((.....))..)))....))....... ( -11.09, z-score = 0.34, R) >consensus GAAAUUUGUUUAUGGACAACUAUUAACAUUUUCCAAUUACACAACAUAACGCUCAUAAAUUAUGCCAUGCAUGUGUAAUUGUUUCGAGCC_CCUCUUCUC__________ ((((..((((.((((....)))).)))).))))((((((((...................(((((...)))))))))))))............................. (-11.32 = -11.26 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:47 2011