| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,196,924 – 9,197,018 |
| Length | 94 |
| Max. P | 0.974499 |

| Location | 9,196,924 – 9,197,018 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.61 |
| Shannon entropy | 0.46097 |
| G+C content | 0.38608 |
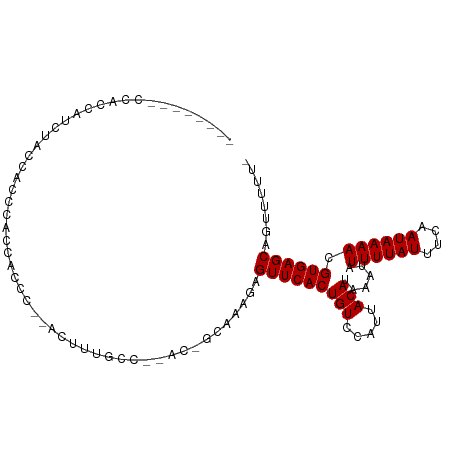
| Mean single sequence MFE | -12.13 |
| Consensus MFE | -7.53 |
| Energy contribution | -7.53 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9196924 94 - 21146708 -------UCCAGCAUCUACCACCCAACACCC--ACUUUGCC--AC-GCAAAGAGUUCACUGUCCAUUACAUAAAUAUUUUAUUUCAAUAAAACGUGAGCAGUUUUU- -------.................(((....--.((((((.--..-)))))).(((((((((.....)))......((((((....)))))).)))))).)))...- ( -13.20, z-score = -2.86, R) >droSim1.chr2R 7656969 94 - 19596830 -------UCCAAAAUCUACCGGCCGCCACCC--ACUUUGCC--AC-GCAAAGAGUUCACUGUCCAUUACAUAAAUAUUUUAUUUCAAUAAAACGUGAGCAGUUUUU- -------.............((......)).--.((((((.--..-)))))).(((((((((.....)))......((((((....)))))).)))))).......- ( -13.00, z-score = -1.22, R) >droSec1.super_1 6719945 94 - 14215200 -------UCCAACAUCUACCACCCGCCACCC--ACUUUGCC--AC-GCAAAGAGUUCACUGUCCAUUACAUAAAUAUUUUAUUUCAAUAAAACGUGAGCAGUUUUU- -------...(((..................--.((((((.--..-)))))).(((((((((.....)))......((((((....)))))).)))))).)))...- ( -13.20, z-score = -2.96, R) >droYak2.chr2R 13376487 101 + 21139217 CCUAAAGCACACCGCCCGCCACCCACCACCC--ACUUUGCC--AC-GCAAAGAGUUCACUGUCCAUUACAUAAAUAUUUUAUUUCAAUAAAACGUGAGCAGUUUUU- ...(((((.......................--.((((((.--..-)))))).(((((((((.....)))......((((((....)))))).)))))).))))).- ( -14.90, z-score = -2.64, R) >droEre2.scaffold_4845 5971388 91 + 22589142 ----------ACCACCCACCACCCACCACCC--AGUUUGCC--AC-GCAAAGAGUUCACUGUCCAUUACAUAAAUAUUUUAUUUCAAUAAAACGUGAGCAGUUUUU- ----------.....................--..(((((.--..-)))))..(((((((((.....)))......((((((....)))))).)))))).......- ( -9.70, z-score = -1.57, R) >droAna3.scaffold_13266 16504740 72 - 19884421 -----------------------------CC--ACUCUGCC--AC-GCAAAGAGUUCACUGUCCAUUACAUAAAUAUUUUAUUUCAAUAAAACGUGAGCAGUUUUU- -----------------------------..--...(((((--((-(.....((....))................((((((....)))))))))).)))).....- ( -9.90, z-score = -1.44, R) >dp4.chr3 19060686 92 + 19779522 ------CCCCCCCAUGUUCCGAUUCCAGC-----CCCAGCC--AC-GCAAACAGUUCACUGUCCAUUACAUAAAUAUUUUAUUUCAAUAAAACGUGAGCAGUUUUU- ------.......................-----....(((--((-(...((((....))))..............((((((....)))))))))).)).......- ( -10.50, z-score = -2.05, R) >droPer1.super_2 8148652 88 + 9036312 ----------CCCAUGUUCCGAUUCCAGC-----CCCAGCC--AC-GCAAAAAGUUCACUGUCCAUUACAUAAAUAUUUUAUUUCAAUAAAACGUGAGCAGUUUUU- ----------....(((((((........-----..(((..--((-.......))...)))...............((((((....)))))))).)))))......- ( -7.80, z-score = -0.43, R) >droWil1.scaffold_181009 2047886 99 - 3585778 -------UCUAUGAUUCGCCUCAUACCCCUCAUACUUCUCCUUAC-GCAAACAGUUCACUGUCCAUUACAUAAAUAUUUUAUUUCAAUAAAACGUGAGCUGCUUUUU -------..(((((......)))))....................-.....(((((((((((.....)))......((((((....)))))).))))))))...... ( -12.70, z-score = -2.83, R) >droGri2.scaffold_15245 10693793 83 - 18325388 -----------------AACAAGUGCGGC----AAGCAGCA--ACAACAAACUGUUCACUGUCCAUUACAUAAAUAUUUUAUUUCAAUAAAACGUGAGCAGUUUUU- -----------------......(((...----..)))...--.....((((((((((((((.....)))......((((((....)))))).)))))))))))..- ( -16.40, z-score = -1.93, R) >consensus ________CCACCAUCUACCACCCACCACCC__ACUUUGCC__AC_GCAAAGAGUUCACUGUCCAUUACAUAAAUAUUUUAUUUCAAUAAAACGUGAGCAGUUUUU_ .....................................................(((((((((.....)))......((((((....)))))).))))))........ ( -7.53 = -7.53 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:42 2011