| Sequence ID | dm3.chr2R |
|---|---|
| Location | 9,112,740 – 9,112,863 |
| Length | 123 |
| Max. P | 0.964260 |

| Location | 9,112,740 – 9,112,849 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.04 |
| Shannon entropy | 0.51562 |
| G+C content | 0.43045 |
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -8.51 |
| Energy contribution | -9.77 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.552532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9112740 109 + 21146708 CUCCAUUCCAUUCUCUUCACUUCGCUGCGAUGAAUUCACCGUAUAUACGGCGAA--AA-AACGCAUCGUUGAUAGUACCACAC--------AUACAUUCAGUUACUCGCUAACACUCUGA ..................(((...(.((((((..(((.((((....)))).)))--..-....)))))).)..))).......--------.........((((.....))))....... ( -16.50, z-score = -0.65, R) >droEre2.scaffold_4845 5884542 113 - 22589142 -----CUCCAUUCUCUUCACUUCGCUGCGAUGAAAUCGCCGUGGAUACGGCGAA--AACAACGCAUCGUUGAUAGUACCACACACACGCACACAUACCCAGUUACUCACUAACACUUCGA -----................(((.((((.((...(((((((....))))))).--..(((((...))))).............))))))..........((((.....))))....))) ( -21.60, z-score = -1.08, R) >droYak2.chr2R 13289276 105 - 21139217 -----CUCCAUUCUCUUCACUUCGCUGCGUUGAAUUCGCCGUGGAAACGGCGAA--AACAACGCAUCGUUGAUAGUACUACAC--------ACAUACUUAGUUGCUCACUAACACUCCAA -----...........(((...((.(((((((..((((((((....))))))))--..))))))).)).)))...........--------......(((((.....)))))........ ( -29.80, z-score = -4.46, R) >droSec1.super_1 6635543 96 + 14215200 -----CUCCAUUCUCUUCACUUCGCUGCGAUGAAUUCACCGUAUAUACGGCGAA--AA-AACGCAUCGUUGAUAGUACCACAC--------ACACAUUUCGUUACUCGCUAA-------- -----.............(((...(.((((((..(((.((((....)))).)))--..-....)))))).)..))).......--------.....................-------- ( -15.60, z-score = -0.74, R) >droSim1.chr2R 7569067 103 + 19596830 -----CUCCAUUCUCUUCACUUCGCUGCGAUGAAUUCACCGUAUAUACGGCGAA--AA-A-CGCAUCGUUGAUAGUACCACAC--------ACACAUUCAGUUACUCGCUAACACUCUGA -----.............(((...(.((((((..(((.((((....)))).)))--..-.-..)))))).)..))).......--------.........((((.....))))....... ( -17.10, z-score = -0.83, R) >droGri2.scaffold_15245 14340419 102 - 18325388 -----CUCGAUUCG-UUCGUUUUAUUGUUGUAAGUCAUCCAUGCAUUCUUUGUUCUAGCAAAACAACAUAAAUAAAACCACGUG-------GUUCAGUCUUUUUCUU-CUAUUAAU---- -----...((..((-(..(((((((((((((..((.......))....(((((....)))))))))))...))))))).)))..-------..))............-........---- ( -12.80, z-score = -1.04, R) >consensus _____CUCCAUUCUCUUCACUUCGCUGCGAUGAAUUCACCGUAUAUACGGCGAA__AA_AACGCAUCGUUGAUAGUACCACAC________ACACAUUCAGUUACUCGCUAACACUC_GA ................(((...((.((((.....(((.((((....)))).))).......)))).)).)))................................................ ( -8.51 = -9.77 + 1.25)


| Location | 9,112,754 – 9,112,863 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.10 |
| Shannon entropy | 0.59436 |
| G+C content | 0.43414 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -8.51 |
| Energy contribution | -9.77 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9112754 109 + 21146708 CUUCACUUCGCUGCGAUGAAUUCACCGUAUAUACGGCGAA--AA-AACGCAUCGUUGAUAGUACCACACAUAC--------AUUCAGUUACUCGCUAACACUCUGACACCCAUGUGUGUG ....(((...(.((((((..(((.((((....)))).)))--..-....)))))).)..)))....(((((((--------((((((...............)))).....))))))))) ( -26.46, z-score = -2.08, R) >droEre2.scaffold_4845 5884551 116 - 22589142 CUUCACUUCGCUGCGAUGAAAUCGCCGUGGAUACGGCGAA--AACAACGCAUCGUUGAUAGUACCACACACACGCACACAUACCCAGUUACUCACUAACACUUCGACACCCAUGUAUA-- .........(.((((.((...(((((((....))))))).--..)).)))).)(((((.(((...........(....).......((((.....))))))))))))...........-- ( -22.80, z-score = -1.00, R) >droYak2.chr2R 13289285 108 - 21139217 CUUCACUUCGCUGCGUUGAAUUCGCCGUGGAAACGGCGAA--AACAACGCAUCGUUGAUAGUACUACACACAU--------ACUUAGUUGCUCACUAACACUCCAACACCCAUGUGUA-- ..(((...((.(((((((..((((((((....))))))))--..))))))).)).))).........((((((--------.....((((.............))))....)))))).-- ( -34.42, z-score = -4.51, R) >droSec1.super_1 6635552 101 + 14215200 CUUCACUUCGCUGCGAUGAAUUCACCGUAUAUACGGCGAA--AA-AACGCAUCGUUGAUAGUACCACACACAC--------AUUUCGUUACUCGCUAA--------CACCCAUUUGUGUG ....(((...(.((((((..(((.((((....)))).)))--..-....)))))).)..)))....((((((.--------.....((((.....)))--------).......)))))) ( -21.82, z-score = -1.57, R) >droSim1.chr2R 7569076 108 + 19596830 CUUCACUUCGCUGCGAUGAAUUCACCGUAUAUACGGCGAA--AA-A-CGCAUCGUUGAUAGUACCACACACAC--------AUUCAGUUACUCGCUAACACUCUGACACCCAUGUGUGUG ....(((...(.((((((..(((.((((....)))).)))--..-.-..)))))).)..)))....(((((((--------((((((...............)))).....))))))))) ( -28.96, z-score = -2.82, R) >droGri2.scaffold_15245 14340428 93 - 18325388 -UUCGUUUUAUUGUUGUAAGUCAUCCAUGCAUUCUUUGUUCUAGCAAAACAACAUAAAUAAAACCACGUGGUU-----CAGUCUUUUUCUUCUAUUAAU--------------------- -...(((((((((((((..((.......))....(((((....)))))))))))...))))))).........-----.....................--------------------- ( -9.90, z-score = -0.63, R) >consensus CUUCACUUCGCUGCGAUGAAUUCACCGUAUAUACGGCGAA__AA_AACGCAUCGUUGAUAGUACCACACACAC________ACUCAGUUACUCACUAACACUC_GACACCCAUGUGUG__ ..(((...((.((((.....(((.((((....)))).))).......)))).)).))).............................................................. ( -8.51 = -9.77 + 1.25)

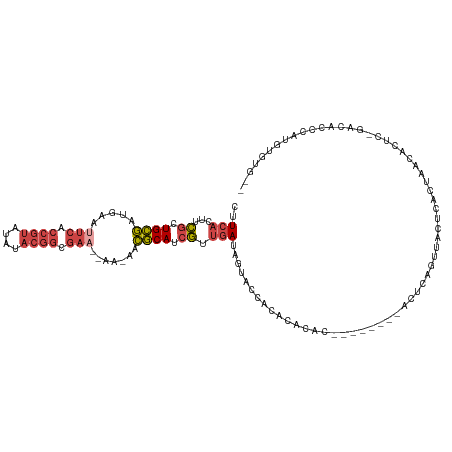
| Location | 9,112,754 – 9,112,863 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.10 |
| Shannon entropy | 0.59436 |
| G+C content | 0.43414 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -16.61 |
| Energy contribution | -17.82 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 9112754 109 - 21146708 CACACACAUGGGUGUCAGAGUGUUAGCGAGUAACUGAAU--------GUAUGUGUGGUACUAUCAACGAUGCGUU-UU--UUCGCCGUAUAUACGGUGAAUUCAUCGCAGCGAAGUGAAG ..((((((((.((.((((..((....)).....))))))--------.))))))))......(((.((.((((..-..--((((((((....)))))))).....)))).))...))).. ( -34.60, z-score = -1.89, R) >droEre2.scaffold_4845 5884551 116 + 22589142 --UAUACAUGGGUGUCGAAGUGUUAGUGAGUAACUGGGUAUGUGUGCGUGUGUGUGGUACUAUCAACGAUGCGUUGUU--UUCGCCGUAUCCACGGCGAUUUCAUCGCAGCGAAGUGAAG --((((((((.(((.((.....(((((.....))))).....))))).))))))))......(((.((.((((.((..--.(((((((....)))))))...)).)))).))...))).. ( -33.80, z-score = -0.11, R) >droYak2.chr2R 13289285 108 + 21139217 --UACACAUGGGUGUUGGAGUGUUAGUGAGCAACUAAGU--------AUGUGUGUAGUACUAUCAACGAUGCGUUGUU--UUCGCCGUUUCCACGGCGAAUUCAACGCAGCGAAGUGAAG --((((((((....((((..((((....)))).))))..--------.))))))))......(((.((.(((((((..--((((((((....))))))))..))))))).))...))).. ( -38.80, z-score = -3.12, R) >droSec1.super_1 6635552 101 - 14215200 CACACAAAUGGGUG--------UUAGCGAGUAACGAAAU--------GUGUGUGUGGUACUAUCAACGAUGCGUU-UU--UUCGCCGUAUAUACGGUGAAUUCAUCGCAGCGAAGUGAAG ((((((.(((..((--------(((.....)))))...)--------)).))))))......(((.((.((((..-..--((((((((....)))))))).....)))).))...))).. ( -30.10, z-score = -1.46, R) >droSim1.chr2R 7569076 108 - 19596830 CACACACAUGGGUGUCAGAGUGUUAGCGAGUAACUGAAU--------GUGUGUGUGGUACUAUCAACGAUGCG-U-UU--UUCGCCGUAUAUACGGUGAAUUCAUCGCAGCGAAGUGAAG ((((((((((....((((..((....)).....)))).)--------)))))))))......(((.((.((((-.-..--((((((((....)))))))).....)))).))...))).. ( -35.60, z-score = -1.88, R) >droGri2.scaffold_15245 14340428 93 + 18325388 ---------------------AUUAAUAGAAGAAAAAGACUG-----AACCACGUGGUUUUAUUUAUGUUGUUUUGCUAGAACAAAGAAUGCAUGGAUGACUUACAACAAUAAAACGAA- ---------------------........(((.........(-----((((....)))))(((((((((..(((((......)))))...))))))))).)))................- ( -13.70, z-score = -0.76, R) >consensus __CACACAUGGGUGUC_GAGUGUUAGCGAGUAACUAAAU________GUGUGUGUGGUACUAUCAACGAUGCGUU_UU__UUCGCCGUAUACACGGUGAAUUCAUCGCAGCGAAGUGAAG ..((((((((............((((.......))))...........))))))))......(((.((.((((.......((((((((....)))))))).....)))).))...))).. (-16.61 = -17.82 + 1.20)

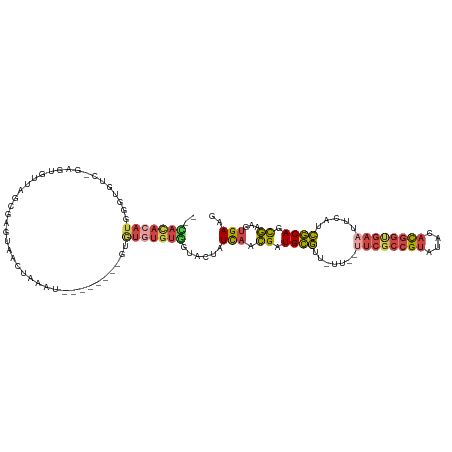
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:28 2011