
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,984,395 – 8,984,531 |
| Length | 136 |
| Max. P | 0.820522 |

| Location | 8,984,395 – 8,984,506 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 88.70 |
| Shannon entropy | 0.15752 |
| G+C content | 0.33709 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -15.39 |
| Energy contribution | -15.83 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8984395 111 - 21146708 ----GUCCUCUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUUGGAACUUGUCUAUACAUAAUUUCUCUACAUUGACUUACUAAUCUAUAAGAUUUUCCAAAACUUUUAUAAAA ----......((((....))))(((((....)))))....((((((((...(((.....................))).....((((.....))))))))))))........... ( -19.20, z-score = -2.10, R) >droSec1.super_1 6510805 115 - 14215200 AUGGGUCGUCAGCUGGAAUGCAGCGACAUCUGUUGCUCAUUUUAGGAACUCGUCCAUACAUAAUUUCUCUACAUUGACAUACUAAUGUAGAAUAUUUUCCAAAACUUUCAAAAAU (((((.((....((((((((.((((((....)))))))))))))).....)))))))..........(((((((((......)))))))))........................ ( -29.90, z-score = -3.75, R) >droSim1.chr2R 7434119 115 - 19596830 AUGGGUUCACUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUAGGAACUCGUCCAUACAUAAUUUCUCUACAUUGACAUACUAAUGUAUAAUAUUUUCCAAAACUUUAAAAAAA ((((((((.(((((....)))((((((....))))))......))))))))))................(((((((......))))))).......................... ( -23.80, z-score = -2.60, R) >consensus AUGGGUCCUCUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUAGGAACUCGUCCAUACAUAAUUUCUCUACAUUGACAUACUAAUGUAUAAUAUUUUCCAAAACUUUAAAAAAA .............(((((((.((((((....)))))).......(((.....)))..............(((((((......))))))).....))))))).............. (-15.39 = -15.83 + 0.45)

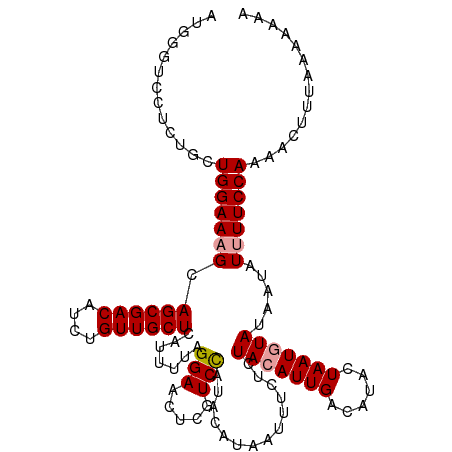

| Location | 8,984,435 – 8,984,531 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 54.16 |
| Shannon entropy | 0.82039 |
| G+C content | 0.37268 |
| Mean single sequence MFE | -21.22 |
| Consensus MFE | -6.80 |
| Energy contribution | -6.74 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.69 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.523468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8984435 96 - 21146708 AACAAAAACUGUGAAAUUUAAGGAC-------------------GUCCUCUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUUGGAACUUGUCUAUACAUAAUUUCUCUACAUU ..........(.((((((((.((((-------------------((((..((((....))))(((((....)))))........)))...))))).))...)))))).)...... ( -20.50, z-score = -1.84, R) >droAna3.scaffold_13266 13437250 93 + 19884421 AUUAAAAAUAGAGCUACUUUGUGGUAAUUAUUUGGAUGAUAGUAAGAUUUUGAUGGCUUGAAAC--CCUCUAAUACAUAACUUUAAA--CAGUCAAU------------------ ........(((((.((((....))))((((((.....)))))).....................--.)))))...............--........------------------ ( -7.20, z-score = 2.34, R) >droYak2.chr2R 13162035 95 + 21139217 AACCGAAAUCAUCAGAUUAAAGGUC------CCCUGCAAAUUGGGAUUUACACUAUGAAGCAGCGACAUCCGUUGCAUAUCUUUGGAACUAGUACGUAAGU-------------- ..(((((....(((......(((((------((.........)))))))......))).((((((.....)))))).....))))).......((....))-------------- ( -21.60, z-score = -1.25, R) >droSec1.super_1 6510845 115 - 14215200 AACAAAACCUUUGAGAUUCAAGGACUACAACUCCUGCAAGAUGGGUCGUCAGCUGGAAUGCAGCGACAUCUGUUGCUCAUUUUAGGAACUCGUCCAUACAUAAUUUCUCUACAUU ............((((....((((.......))))....(((((((..((..((((((((.((((((....)))))))))))))))))))))))...........))))...... ( -28.40, z-score = -1.78, R) >droSim1.chr2R 7434159 115 - 19596830 AACAAAAACUGUGAGAUUUAAGGACUACAACUCCUGCAAAAUGGGUUCACUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUAGGAACUCGUCCAUACAUAAUUUCUCUACAUU .........(((((((.(((.((((......(((((..((((((((((.(((((....))))).))........)))))))))))))....)))).....)))..)))).))).. ( -28.40, z-score = -2.49, R) >consensus AACAAAAACUGUGAGAUUUAAGGAC_A__A_UCCUGCAAAAUGGGUCCUCUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUUGGAACUAGUCCAUACAUAAUUUCUCUACAUU .....................((((......(((........)))..............((((((.....))))))...............)))).................... ( -6.80 = -6.74 + -0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:18 2011