| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,960,057 – 8,960,151 |
| Length | 94 |
| Max. P | 0.920815 |

| Location | 8,960,057 – 8,960,151 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.63 |
| Shannon entropy | 0.55609 |
| G+C content | 0.49081 |
| Mean single sequence MFE | -32.26 |
| Consensus MFE | -14.91 |
| Energy contribution | -15.17 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
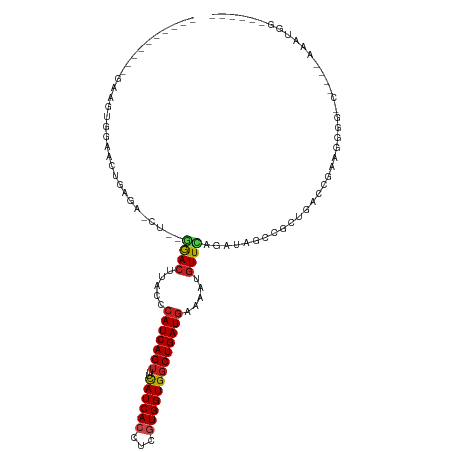
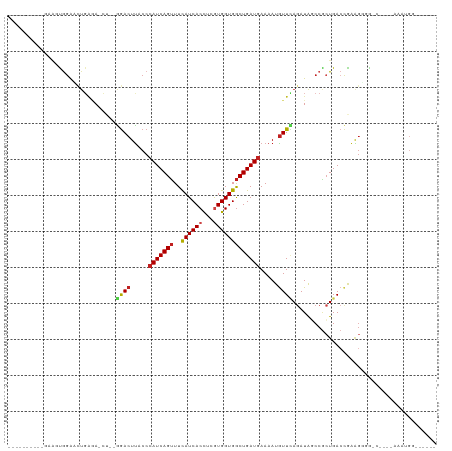
>dm3.chr2R 8960057 94 + 21146708 ----------GAAGCGGAACUGAGA-CU--GGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACCGAAGUGG------AAAUGG------ ----------.....((((((((..-.(--((......))))))))))).((((.(((.((((((((.((((....))))..))))))))...))).))))------......------ ( -31.90, z-score = -2.69, R) >droSim1.chr2R 7408564 94 + 19596830 ----------GAAGCGGAACUGAGA-CU--GGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACCGAAGUGG------AAAUGG------ ----------.....((((((((..-.(--((......))))))))))).((((.(((.((((((((.((((....))))..))))))))...))).))))------......------ ( -31.90, z-score = -2.69, R) >droSec1.super_1 6476383 94 + 14215200 ----------GAAGCGGAACUGAAA-CU--GGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACCGAAGUGG------AAAUGG------ ----------.....((((((((..-.(--((......))))))))))).((((.(((.((((((((.((((....))))..))))))))...))).))))------......------ ( -31.90, z-score = -3.06, R) >droYak2.chr2R 7962076 94 + 21139217 ----------CAAGCGGAACUGAGA-CU--GGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACCGAAGUGG------AAAUGG------ ----------.....((((((((..-.(--((......))))))))))).((((.(((.((((((((.((((....))))..))))))))...))).))))------......------ ( -31.90, z-score = -2.62, R) >droEre2.scaffold_4845 5731252 93 - 22589142 -----------AAGCGGAAAUGAGA-CU--GGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACCGAGGUGG------ACAUGG------ -----------.........(((((-((--((.........)))))))))((((((((.((((((((.((((....))))..))))))))...))))))))------......------ ( -34.70, z-score = -3.13, R) >droAna3.scaffold_13266 6429884 106 + 19884421 ----------GAAAUGAGACUGGAA-CU--GGACUCACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACGGAGGGCGUCCGCCAAGUGGAAAUGG ----------...((((((((((..-.(--((......)))))))))))))..((((((((((((((.((((....))))..)))))))).)).))))...(((((...)))))..... ( -38.00, z-score = -2.09, R) >dp4.chr3 5730460 117 + 19779522 AGGGGGACUGUAACUGGAACUGAGAACA--GGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUAUUCAGAUAUCCUCUGACCAAAGGCGUCCGCCAAGAGGCCAACG .(..((........(((((((((.....--((......)).)))))))))...((((....((((.((((.......(((((.....))))).......)))).)))).))))))..). ( -35.84, z-score = -1.05, R) >droPer1.super_2 3062248 117 - 9036312 UAGGGGACUGGAACUGGAACUGAGAACA--GGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUGGCCGCUGACCAAAGGCGUCCGCCAAGUGGCCAACG (((....)))(((((((((((((.....--((......)).)))))))))(((((....)))))...........)))).(.((((((((.......(((....))).)))))))).). ( -40.50, z-score = -1.43, R) >droWil1.scaffold_180700 314246 95 + 6630534 --------GUCUGGUGGGUGGGGGAGGGA-AAACUUACCCAUCAGUUUCAUCACUUCAUGGUGGCUGAUGAAAAUGUUUAAAUAGCCGCUGACUGGAAGGGCCA--------------- --------..((((((((((((.......-...))))))))))))........((((.(((((((((.((((....))))..)))))))))....)))).....--------------- ( -32.00, z-score = -1.46, R) >droVir3.scaffold_12875 719299 107 + 20611582 ----------GAAAUAGGUUUCAUAUGU--GGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUAAAAAUGUUUAGAUAUCCGCUGACCAAAGGUAGCUGCCAAGUGAUCCAAG ----------......((..((((...(--((..(((((.((((((..((((((...)))))))))))).....((.((((.......)))).))..)))))...))).)))).))... ( -25.80, z-score = -0.83, R) >droMoj3.scaffold_6496 3735391 109 + 26866924 ----------AAUAUGGGACUCAUAUGUAUAGACUUACCCAUCAGCUUCAUCACUUCGUGGUGACUGAUGAAAAUGUUGAGAUAUCCGCUGACCGGAGGCAGCUGCCAGGAGAGCAGCG ----------......(((((((...(((......))).((((((..(((((((...))))))))))))).......))))...)))((((.((...(((....))).))....)))). ( -31.90, z-score = -0.92, R) >droGri2.scaffold_15245 10188777 92 + 18325388 ----------GACAUAGGUCUGUGAUGU--AGACUUACCCAUCAGUUUUAUCACUUCAUGGUGGCUGAUGAAAAUGUUGAGAUAUCCAGUGAGCAGUGACAAGU--------------- ----------..(((.(.((((.(((((--.(((.....(((((((.....((((....))))))))))).....)))...)))))))..)).).)))......--------------- ( -20.80, z-score = 0.35, R) >consensus __________GAAGUGGAACUGAGA_CU__GGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACCGAAGGGG_C____AAAUGG______ ..............................((((.....(((((((..((((((...))))))))))))).....))))........................................ (-14.91 = -15.17 + 0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:16 2011