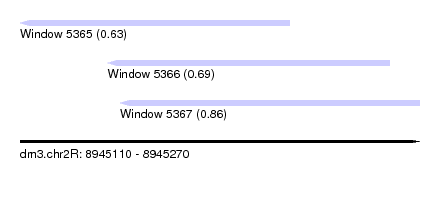
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 8,945,110 – 8,945,270 |
| Length | 160 |
| Max. P | 0.861175 |

| Location | 8,945,110 – 8,945,218 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.53 |
| Shannon entropy | 0.41591 |
| G+C content | 0.46476 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -19.00 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8945110 108 - 21146708 AUGCUUUAAGGCCACACGGCGUAUGAGUGAUGCAAUUUUCUGGCCUUAAUGUUGAAUCUUUCCUCGAACAAAUGGC------------CACACUGCGUAUGCGUAAUGCUUUAGACCAAC .((.((((((((...(((.((((((((((...........(((((......((((........))))......)))------------)))))).)))))))))...)))))))).)).. ( -30.20, z-score = -1.86, R) >droSim1.chr2R 7397328 108 - 19596830 AUGCUUUAAGGCCACACGGCGUAUGAUUGAUGCCAUUUUCUGGCCUUAAAGUUCAAUCCCACUUCGAACAUAGUGCAAGUAAGG---CCACACUGCGUAUGCGUGAUGCAC--------- ..((((((((((((...(((((.......)))))......))))))))))))...........(((..((((.((((.((....---....))))))))))..))).....--------- ( -29.90, z-score = -0.87, R) >droSec1.super_1 6461403 106 - 14215200 AUGCUUUAAGGCCACACGGCGUAUGAGUGAUGCCAUUUUCUGGCCUUAAAGUUCAAUCCCACUUCGAACAUAGUGCAAGUAAGG---CCACACUGCGUAUACGUGAUUC----------- ..((((((((((((...(((((.......)))))......))))))))))))(((....((((........))))...((((.(---(......)).).))).)))...----------- ( -27.50, z-score = -0.92, R) >droYak2.chr2R 13121748 109 + 21139217 AUACUUUAAGGCUACACUGCGUAUGAGCAAUGUAGUUUGCUGGCCUUAAAGUUCCAUCUCACUUCGAACAUAGAGCAACUAAGGGCAACACACUGCGUAUGCGUGAUGC----------- ..((((((((((((.(((((((.......)))))))....))))))))))))...........(((..((((..(((......(....)....))).))))..)))...----------- ( -28.60, z-score = -0.39, R) >droEre2.scaffold_4845 5716610 107 + 22589142 AUACUUUAGGGCCACACGGCGUAUGAGUGAUGC-ACUGGCUGGCCUUAAAGUUCAAUCUUACUUCGAACACAGUGCCACUAAGG-CCACACUUUUCACCAACGCACUGC----------- ..((((((((((((..(((.((((.....))))-.)))..))))))))))))..................((((((....(((.-.....))).........)))))).----------- ( -32.32, z-score = -2.42, R) >consensus AUGCUUUAAGGCCACACGGCGUAUGAGUGAUGCAAUUUUCUGGCCUUAAAGUUCAAUCUCACUUCGAACAUAGUGCAACUAAGG___ACACACUGCGUAUGCGUGAUGC___________ ..((((((((((((...(((((.......)))))......)))))))))))).........................................((((....))))............... (-19.00 = -18.88 + -0.12)

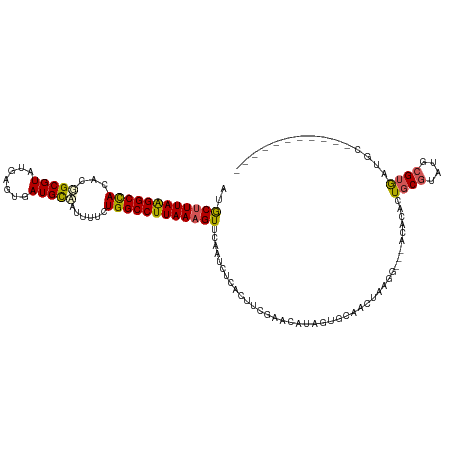
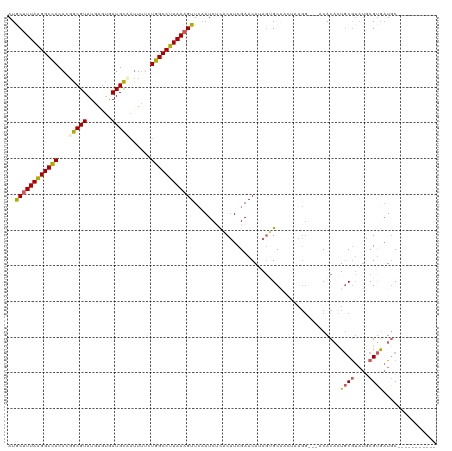
| Location | 8,945,145 – 8,945,258 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 78.80 |
| Shannon entropy | 0.37959 |
| G+C content | 0.44823 |
| Mean single sequence MFE | -30.41 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.30 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.687374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8945145 113 - 21146708 GCGAUGUGUAUACCAGCUA-GUGCGCACAAAUGAACAUCGAAUGCUUUAAGGCCACACGGCGUAUGAGUGAUGCAAUUUUCUGGCCUUAAUGUUGAAUCUUUCCUCGAACAAAU------- .(((((((..(((......-)))..))))........((((.....(((((((((....((((.......)))).......)))))))))..))))........))).......------- ( -25.70, z-score = 0.11, R) >droSim1.chr2R 7397356 120 - 19596830 GCGAUAUGCAUACCAGCUA-GUCCGCACAAAUGAACAUCGAAUGCUUUAAGGCCACACGGCGUAUGAUUGAUGCCAUUUUCUGGCCUUAAAGUUCAAUCCCACUUCGAACAUAGUGCAAGU (.(((..((......))..-))))((((..(((....(((((.((((((((((((...(((((.......)))))......))))))))))))..........))))).))).)))).... ( -32.20, z-score = -1.96, R) >droSec1.super_1 6461429 120 - 14215200 GCGAUAUGUAUACCAGCUA-GUGCGCACAAAUGAACAUCGAAUGCUUUAAGGCCACACGGCGUAUGAGUGAUGCCAUUUUCUGGCCUUAAAGUUCAAUCCCACUUCGAACAUAGUGCAAGU (((.(((((......((..-..)).............(((((.((((((((((((...(((((.......)))))......))))))))))))..........)))))))))).))).... ( -33.90, z-score = -1.82, R) >droYak2.chr2R 13121777 114 + 21139217 ------GCUAUUUCAGUUA-GAGCGCACAAAUGAACAUCGAAUACUUUAAGGCUACACUGCGUAUGAGCAAUGUAGUUUGCUGGCCUUAAAGUUCCAUCUCACUUCGAACAUAGAGCAACU ------(((..((((....-...........))))..(((((.((((((((((((.(((((((.......)))))))....))))))))))))..........)))))......))).... ( -26.06, z-score = -0.68, R) >droEre2.scaffold_4845 5716638 105 + 22589142 ---------------GCGAUGUGCGCGCAAAUGAACAUCGAAUACUUUAGGGCCACACGGCGUAUGAGUGAUGC-ACUGGCUGGCCUUAAAGUUCAAUCUUACUUCGAACACAGUGCCACU ---------------.....(((.((((...((....(((((.((((((((((((..(((.((((.....))))-.)))..))))))))))))..........))))).))..))))))). ( -34.20, z-score = -2.13, R) >consensus GCGAU_UGUAUACCAGCUA_GUGCGCACAAAUGAACAUCGAAUGCUUUAAGGCCACACGGCGUAUGAGUGAUGCAAUUUUCUGGCCUUAAAGUUCAAUCUCACUUCGAACAUAGUGCAACU ...............((.......))...........(((((.((((((((((((...(((((.......)))))......))))))))))))..........)))))............. (-21.54 = -21.30 + -0.24)
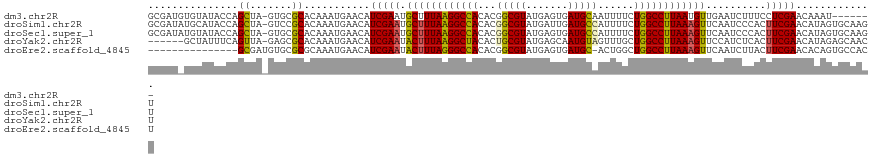
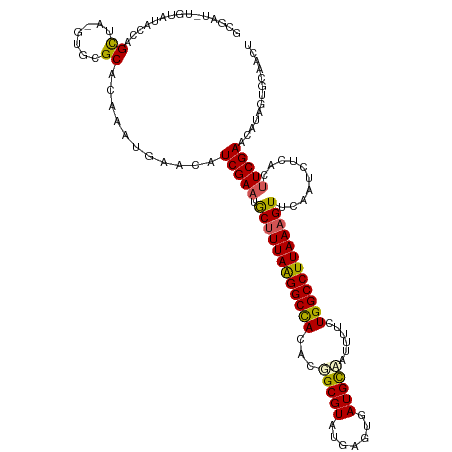
| Location | 8,945,150 – 8,945,270 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Shannon entropy | 0.30900 |
| G+C content | 0.45612 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -27.70 |
| Energy contribution | -28.70 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 8945150 120 - 21146708 CACUGCGUAUGAGCGAUGUGUAUACCAGCUAGUGCGCACAAAUGAACAUCGAAUGCUUUAAGGCCACACGGCGUAUGAGUGAUGCAAUUUUCUGGCCUUAAUGUUGAAUCUUUCCUCGAA ...(((((((.(((...((....))..))).)))))))..........((((..((.(((((((((....((((.......)))).......))))))))).))...........)))). ( -29.52, z-score = 0.20, R) >droSim1.chr2R 7397368 120 - 19596830 CGCUGCGUAUGAGCGAUAUGCAUACCAGCUAGUCCGCACAAAUGAACAUCGAAUGCUUUAAGGCCACACGGCGUAUGAUUGAUGCCAUUUUCUGGCCUUAAAGUUCAAUCCCACUUCGAA .((((.(((((.........)))))))))...................(((((.((((((((((((...(((((.......)))))......))))))))))))..........))))). ( -35.00, z-score = -2.18, R) >droSec1.super_1 6461441 120 - 14215200 CGCUGCGUUUGAGCGAUAUGUAUACCAGCUAGUGCGCACAAAUGAACAUCGAAUGCUUUAAGGCCACACGGCGUAUGAGUGAUGCCAUUUUCUGGCCUUAAAGUUCAAUCCCACUUCGAA .(.(((((...(((.............)))...)))))).........(((((.((((((((((((...(((((.......)))))......))))))))))))..........))))). ( -34.92, z-score = -1.61, R) >droYak2.chr2R 13121789 114 + 21139217 CACGGCGUAUAAGCUAUUU------CAGUUAGAGCGCACAAAUGAACAUCGAAUACUUUAAGGCUACACUGCGUAUGAGCAAUGUAGUUUGCUGGCCUUAAAGUUCCAUCUCACUUCGAA ....((((.(.((((....------.)))).).))))...........(((((.((((((((((((.(((((((.......)))))))....))))))))))))..........))))). ( -29.60, z-score = -1.39, R) >droEre2.scaffold_4845 5716650 105 + 22589142 CAUUGCGUAUGAGCGAU--------------GUGCGCGCAAAUGAACAUCGAAUACUUUAGGGCCACACGGCGUAUGAGUGAUGCA-CUGGCUGGCCUUAAAGUUCAAUCUUACUUCGAA ....((((((.......--------------))))))...........(((((.((((((((((((..(((.((((.....)))).-)))..))))))))))))..........))))). ( -34.60, z-score = -2.11, R) >consensus CACUGCGUAUGAGCGAUAUG_AUACCAGCUAGUGCGCACAAAUGAACAUCGAAUGCUUUAAGGCCACACGGCGUAUGAGUGAUGCAAUUUUCUGGCCUUAAAGUUCAAUCUCACUUCGAA ....((((((.(((.............))).))))))...........(((((.((((((((((((...(((((.......)))))......))))))))))))..........))))). (-27.70 = -28.70 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:19:14 2011